Discovery and functional characterization of two diterpene synthases for sclareol biosynthesis in Salvia sclarea (L.) and their relevance for perfume manufacture
- PMID: 22834731
- PMCID: PMC3520730
- DOI: 10.1186/1471-2229-12-119
Discovery and functional characterization of two diterpene synthases for sclareol biosynthesis in Salvia sclarea (L.) and their relevance for perfume manufacture
Abstract
Background: Sclareol is a diterpene natural product of high value for the fragrance industry. Its labdane carbon skeleton and its two hydroxyl groups also make it a valued starting material for semisynthesis of numerous commercial substances, including production of Ambrox® and related ambergris substitutes used in the formulation of high end perfumes. Most of the commercially-produced sclareol is derived from cultivated clary sage (Salvia sclarea) and extraction of the plant material. In clary sage, sclareol mainly accumulates in essential oil-producing trichomes that densely cover flower calices. Manool also is a minor diterpene of this species and the main diterpene of related Salvia species.
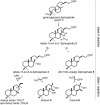
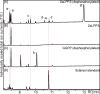
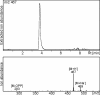
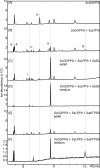
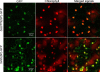
Results: Based on previous general knowledge of diterpene biosynthesis in angiosperms, and based on mining of our recently published transcriptome database obtained by deep 454-sequencing of cDNA from clary sage calices, we cloned and functionally characterized two new diterpene synthase (diTPS) enzymes for the complete biosynthesis of sclareol in clary sage. A class II diTPS (SsLPPS) produced labda-13-en-8-ol diphosphate as major product from geranylgeranyl diphosphate (GGPP) with some minor quantities of its non-hydroxylated analogue, (9 S, 10 S)-copalyl diphosphate. A class I diTPS (SsSS) then transformed these intermediates into sclareol and manool, respectively. The production of sclareol was reconstructed in vitro by combining the two recombinant diTPS enzymes with the GGPP starting substrate and in vivo by co-expression of the two proteins in yeast (Saccharomyces cerevisiae). Tobacco-based transient expression assays of green fluorescent protein-fusion constructs revealed that both enzymes possess an N-terminal signal sequence that actively targets SsLPPS and SsSS to the chloroplast, a major site of GGPP and diterpene production in plants.
Conclusions: SsLPPS and SsSS are two monofunctional diTPSs which, together, produce the diterpenoid specialized metabolite sclareol in a two-step process. They represent two of the first characterized hydroxylating diTPSs in angiosperms and generate the dihydroxylated labdane sclareol without requirement for additional enzymatic oxidation by activities such as cytochrome P450 monoxygenases. Yeast-based production of sclareol by co-expresssion of SsLPPS and SsSS was efficient enough to warrant the development and use of such technology for the biotechnological production of scareol and other oxygenated diterpenes.
Figures

Similar articles
-
Evolution of conifer diterpene synthases: diterpene resin acid biosynthesis in lodgepole pine and jack pine involves monofunctional and bifunctional diterpene synthases.Plant Physiol. 2013 Feb;161(2):600-16. doi: 10.1104/pp.112.208546. Epub 2012 Dec 12. Plant Physiol. 2013. PMID: 23370714 Free PMC article.
-
Enzymes for synthetic biology of ambroxide-related diterpenoid fragrance compounds.Adv Biochem Eng Biotechnol. 2015;148:427-47. doi: 10.1007/10_2015_308. Adv Biochem Eng Biotechnol. 2015. PMID: 25846965 Review.
-
A diterpene synthase from the clary sage Salvia sclarea catalyzes the cyclization of geranylgeranyl diphosphate to (8R)-hydroxy-copalyl diphosphate.Phytochemistry. 2013 Jul;91:93-9. doi: 10.1016/j.phytochem.2012.07.019. Epub 2012 Sep 6. Phytochemistry. 2013. PMID: 22959531
-
Biosynthesis of the psychotropic plant diterpene salvinorin A: Discovery and characterization of the Salvia divinorum clerodienyl diphosphate synthase.Plant J. 2017 Mar;89(5):885-897. doi: 10.1111/tpj.13427. Epub 2017 Feb 6. Plant J. 2017. PMID: 27865008
-
Plant diterpene synthases: exploring modularity and metabolic diversity for bioengineering.Trends Biotechnol. 2015 Jul;33(7):419-28. doi: 10.1016/j.tibtech.2015.04.006. Epub 2015 May 20. Trends Biotechnol. 2015. PMID: 26003209 Review.
Cited by
-
Evolution of conifer diterpene synthases: diterpene resin acid biosynthesis in lodgepole pine and jack pine involves monofunctional and bifunctional diterpene synthases.Plant Physiol. 2013 Feb;161(2):600-16. doi: 10.1104/pp.112.208546. Epub 2012 Dec 12. Plant Physiol. 2013. PMID: 23370714 Free PMC article.
-
Genome-wide detection of terpene synthase genes in holy basil (Ocimum sanctum L.).PLoS One. 2018 Nov 16;13(11):e0207097. doi: 10.1371/journal.pone.0207097. eCollection 2018. PLoS One. 2018. PMID: 30444870 Free PMC article.
-
Functional divergence of CYP76AKs shapes the chemodiversity of abietane-type diterpenoids in genus Salvia.Nat Commun. 2023 Aug 4;14(1):4696. doi: 10.1038/s41467-023-40401-y. Nat Commun. 2023. PMID: 37542034 Free PMC article.
-
Manoyl oxide (13R), the biosynthetic precursor of forskolin, is synthesized in specialized root cork cells in Coleus forskohlii.Plant Physiol. 2014 Mar;164(3):1222-36. doi: 10.1104/pp.113.228429. Epub 2014 Jan 30. Plant Physiol. 2014. PMID: 24481136 Free PMC article.
-
Biochemical characterization of the castor bean ent-kaurene synthase(-like) family supports quantum chemical view of diterpene cyclization.Phytochemistry. 2014 Jul;103:13-21. doi: 10.1016/j.phytochem.2014.04.005. Epub 2014 May 5. Phytochemistry. 2014. PMID: 24810014 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

