NF45 dimerizes with NF90, Zfr and SPNR via a conserved domain that has a nucleotidyltransferase fold
- PMID: 22833610
- PMCID: PMC3467086
- DOI: 10.1093/nar/gks696
NF45 dimerizes with NF90, Zfr and SPNR via a conserved domain that has a nucleotidyltransferase fold
Abstract
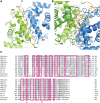
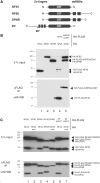
Nuclear factors NF90 and NF45 form a complex involved in a variety of cellular processes and are thought to affect gene expression both at the transcriptional and translational level. In addition, this complex affects the replication of several viruses through direct interactions with viral RNA. NF90 and NF45 dimerize through their common 'DZF' domain (domain associated with zinc fingers). NF90 has additional double-stranded RNA-binding domains that likely mediate its association with target RNAs. We present the crystal structure of the NF90/NF45 dimerization complex at 1.9-Å resolution. The DZF domain shows structural similarity to the template-free nucleotidyltransferase family of RNA modifying enzymes. However, both NF90 and NF45 have lost critical catalytic residues during evolution and are therefore not functional enzymes. Residues on NF90 that make up its interface with NF45 are conserved in two related proteins, spermatid perinuclear RNA-binding protein (SPNR) and zinc-finger RNA-binding protein (Zfr). Using a co-immunoprecipitation assay and site-specific mutants, we demonstrate that NF45 is also able to recognize SPNR and Zfr through the same binding interface, revealing that NF45 is able to form a variety of cellular complexes with other DZF-domain proteins.
Figures



Similar articles
-
NF90-NF45 is a selective RNA chaperone that rearranges viral and cellular riboswitches: biochemical analysis of a virus host factor activity.Nucleic Acids Res. 2017 Dec 1;45(21):12441-12454. doi: 10.1093/nar/gkx931. Nucleic Acids Res. 2017. PMID: 29040738 Free PMC article.
-
The properties of the RNA-binding protein NF90 are considerably modulated by complex formation with NF45.Biochem J. 2017 Jan 15;474(2):259-280. doi: 10.1042/BCJ20160790. Epub 2016 Nov 14. Biochem J. 2017. PMID: 28062840
-
NF45 and NF90 Bind HIV-1 RNA and Modulate HIV Gene Expression.Viruses. 2016 Feb 16;8(2):47. doi: 10.3390/v8020047. Viruses. 2016. PMID: 26891316 Free PMC article.
-
NF90 isoforms, a new family of cellular proteins involved in viral replication?Biochimie. 2015 Jan;108:20-4. doi: 10.1016/j.biochi.2014.10.022. Epub 2014 Nov 4. Biochimie. 2015. PMID: 25447144 Review.
-
Ilf3 and NF90 functions in RNA biology.Wiley Interdiscip Rev RNA. 2015 Mar-Apr;6(2):243-56. doi: 10.1002/wrna.1270. Epub 2014 Oct 18. Wiley Interdiscip Rev RNA. 2015. PMID: 25327818 Review.
Cited by
-
Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.Nucleic Acids Res. 2021 May 7;49(8):4725-4737. doi: 10.1093/nar/gkab165. Nucleic Acids Res. 2021. PMID: 33836064 Free PMC article.
-
ILF3 contributes to the establishment of the antiviral type I interferon program.Nucleic Acids Res. 2020 Jan 10;48(1):116-129. doi: 10.1093/nar/gkz1060. Nucleic Acids Res. 2020. PMID: 31701124 Free PMC article.
-
Miocene Diversification and High-Altitude Adaptation of Parnassius Butterflies (Lepidoptera: Papilionidae) in Qinghai-Tibet Plateau Revealed by Large-Scale Transcriptomic Data.Insects. 2020 Nov 3;11(11):754. doi: 10.3390/insects11110754. Insects. 2020. PMID: 33153157 Free PMC article.
-
Knockdown of ZFR suppresses cell proliferation and invasion of human pancreatic cancer.Biol Res. 2016 May 13;49(1):26. doi: 10.1186/s40659-016-0086-3. Biol Res. 2016. PMID: 27177590 Free PMC article.
-
NF90-NF45 is a selective RNA chaperone that rearranges viral and cellular riboswitches: biochemical analysis of a virus host factor activity.Nucleic Acids Res. 2017 Dec 1;45(21):12441-12454. doi: 10.1093/nar/gkx931. Nucleic Acids Res. 2017. PMID: 29040738 Free PMC article.
References
-
- Marcoulatos P, Koussidis G, Mamuris Z, Velissariou V, Vamvakopoulos NC. Mapping interleukin enhancer binding factor 2 gene (ILF2) to human chromosome 1 (1q11-qter and 1p11-p12) by polymerase chain reaction amplification of human-rodent somatic cell hybrid DNA templates. J. Interferon Cytokine Res. 1996;16:1035–1038. - PubMed
-
- Barber GN. The NFAR's (nuclear factors associated with dsRNA): evolutionarily conserved members of the dsRNA binding protein family. RNA Biol. 2009;6:35–39. - PubMed
-
- Corthesy B, Kao PN. Purification by DNA affinity chromatography of two polypeptides that contact the NF-AT DNA binding site in the interleukin 2 promoter. J. Biol. Chem. 1994;269:20682–20690. - PubMed
-
- Kao PN, Chen L, Brock G, Ng J, Kenny J, Smith AJ, Corthesy B. Cloning and expression of cyclosporin A- and FK506-sensitive nuclear factor of activated T-cells: NF45 and NF90. J. Biol. Chem. 1994;269:20691–20699. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

