Complexity and variability of gut commensal microbiota in polyphagous lepidopteran larvae
- PMID: 22815679
- PMCID: PMC3398904
- DOI: 10.1371/journal.pone.0036978
Complexity and variability of gut commensal microbiota in polyphagous lepidopteran larvae
Abstract
Background: The gut of most insects harbours nonpathogenic microorganisms. Recent work suggests that gut microbiota not only provide nutrients, but also involve in the development and maintenance of the host immune system. However, the complexity, dynamics and types of interactions between the insect hosts and their gut microbiota are far from being well understood.
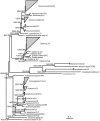
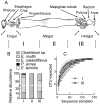
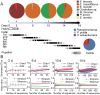
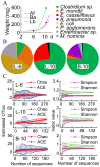
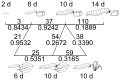
Methods/principal findings: To determine the composition of the gut microbiota of two lepidopteran pests, Spodoptera littoralis and Helicoverpa armigera, we applied cultivation-independent techniques based on 16S rRNA gene sequencing and microarray. The two insect species were very similar regarding high abundant bacterial families. Different bacteria colonize different niches within the gut. A core community, consisting of Enterococci, Lactobacilli, Clostridia, etc. was revealed in the insect larvae. These bacteria are constantly present in the digestion tract at relatively high frequency despite that developmental stage and diet had a great impact on shaping the bacterial communities. Some low-abundant species might become dominant upon loading external disturbances; the core community, however, did not change significantly. Clearly the insect gut selects for particular bacterial phylotypes.
Conclusions: Because of their importance as agricultural pests, phytophagous Lepidopterans are widely used as experimental models in ecological and physiological studies. Our results demonstrated that a core microbial community exists in the insect gut, which may contribute to the host physiology. Host physiology and food, nevertheless, significantly influence some fringe bacterial species in the gut. The gut microbiota might also serve as a reservoir of microorganisms for ever-changing environments. Understanding these interactions might pave the way for developing novel pest control strategies.
Conflict of interest statement
Figures
Similar articles
-
Host plant induced variation in gut bacteria of Helicoverpa armigera.PLoS One. 2012;7(1):e30768. doi: 10.1371/journal.pone.0030768. Epub 2012 Jan 26. PLoS One. 2012. PMID: 22292034 Free PMC article.
-
Census of the bacterial community of the gypsy moth larval midgut by using culturing and culture-independent methods.Appl Environ Microbiol. 2004 Jan;70(1):293-300. doi: 10.1128/AEM.70.1.293-300.2004. Appl Environ Microbiol. 2004. PMID: 14711655 Free PMC article.
-
Comparative evaluation of the gut microbiota associated with the below- and above-ground life stages (larvae and beetles) of the forest cockchafer, Melolontha hippocastani.PLoS One. 2012;7(12):e51557. doi: 10.1371/journal.pone.0051557. Epub 2012 Dec 10. PLoS One. 2012. PMID: 23251574 Free PMC article.
-
Recent advances in molecular approaches to gut microbiota in inflammatory bowel disease.Curr Pharm Des. 2009;15(18):2066-73. doi: 10.2174/138161209788489186. Curr Pharm Des. 2009. PMID: 19519444 Review.
-
Microbial diversity in the human intestine and novel insights from metagenomics.Front Biosci (Landmark Ed). 2009 Jan 1;14(9):3214-21. doi: 10.2741/3445. Front Biosci (Landmark Ed). 2009. PMID: 19273267 Review.
Cited by
-
The Diversity of Wolbachia and Other Bacterial Symbionts in Spodoptera frugiperda.Insects. 2024 Mar 22;15(4):217. doi: 10.3390/insects15040217. Insects. 2024. PMID: 38667347 Free PMC article.
-
The immunostimulatory role of an Enterococcus-dominated gut microbiota in host protection against bacterial and fungal pathogens in Galleria mellonella larvae.Front Insect Sci. 2023 Oct 26;3:1260333. doi: 10.3389/finsc.2023.1260333. eCollection 2023. Front Insect Sci. 2023. PMID: 38469511 Free PMC article.
-
Variation of Helicoverpa armigera symbionts across developmental stages and geographic locations.Front Microbiol. 2023 Sep 7;14:1251627. doi: 10.3389/fmicb.2023.1251627. eCollection 2023. Front Microbiol. 2023. PMID: 37744901 Free PMC article.
-
Impact of gut microbiota composition on black cutworm, Agrotis ipsilon (hufnagel) metabolic indices and pesticide degradation.Anim Microbiome. 2023 Sep 15;5(1):44. doi: 10.1186/s42523-023-00264-6. Anim Microbiome. 2023. PMID: 37715236 Free PMC article.
-
Comparative Genomics of Pesticide-Degrading Enterococcus Symbionts of Spodoptera frugiperda (Lepidoptera: Noctuidae) Leads to the Identification of Two New Species and the Reappraisal of Insect-Associated Enterococcus Species.Microb Ecol. 2023 Nov;86(4):2583-2605. doi: 10.1007/s00248-023-02264-0. Epub 2023 Jul 11. Microb Ecol. 2023. PMID: 37433981
References
-
- Gil R, Latorre A, Moya A. Bacterial endosymbionts of insects: insights from comparative genomics. Environ Microbiol. 2004;6:1109–1122. - PubMed
-
- Wernegreen JJ. Genome evolution in bacterial endosymbionts of insects. Nat Rev Genet. 2002;3:850–861. - PubMed
-
- Brauman A, Kane MD, Labat M, Breznak JA. Genesis of acetate and methane by gut bacteria of nutritionally diverse termites. Science. 1992;257:1384–1387. - PubMed
-
- Chen DQ, Purcell AH. Occurrence and transmission of facultative endosymbionts in aphids. Curr Microbiol. 1997;34:220–225. - PubMed
-
- Warnecke F, Luginbuhl P, Ivanova N, Ghassemian M, Richardson TH, et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature. 2007;450:560–565. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

