Availability of Rubisco small subunit up-regulates the transcript levels of large subunit for stoichiometric assembly of its holoenzyme in rice
- PMID: 22811433
- PMCID: PMC3440226
- DOI: 10.1104/pp.112.201459
Availability of Rubisco small subunit up-regulates the transcript levels of large subunit for stoichiometric assembly of its holoenzyme in rice
Abstract
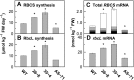
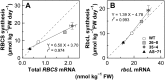
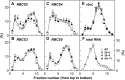
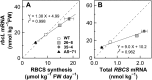
Rubisco is composed of eight small subunits coded for by the nuclear RBCS multigene family and eight large subunits coded for by the rbcL gene in the plastome. For synthesis of the Rubisco holoenzyme, both genes need to be expressed coordinately. To investigate this molecular mechanism, the protein synthesis of two subunits of Rubisco was characterized in transgenic rice (Oryza sativa) plants with overexpression or antisense suppression of the RBCS gene. Total RBCS and rbcL messenger RNA (mRNA) levels and RBCS and RbcL synthesis simultaneously increased in RBCS-sense plants, although the increase in total RBCS mRNA level was greater. In RBCS-antisense plants, the levels of these mRNAs and the synthesis of the corresponding proteins declined to a similar extent. The amount of RBCS synthesized was tightly correlated with rbcL mRNA level among genotypes but not associated with changes in mRNA levels of other major chloroplast-encoded photosynthetic genes. The level of rbcL mRNA, in turn, was tightly correlated with the amount of RbcL synthesized, the molar ratio of RBCS synthesis to RbcL synthesis being identical irrespective of genotype. Polysome loading of rbcL mRNA was not changed. These results demonstrate that the availability of RBCS protein up-regulates the gene expression of rbcL primarily at the transcript level in a quantitative manner for stoichiometric assembly of Rubisco holoenzyme.
Figures

Similar articles
-
Translational downregulation of RBCL is operative in the coordinated expression of Rubisco genes in senescent leaves in rice.J Exp Bot. 2013 Feb;64(4):1145-52. doi: 10.1093/jxb/ers398. Epub 2013 Jan 23. J Exp Bot. 2013. PMID: 23349140 Free PMC article.
-
Effect of individual suppression of RBCS multigene family on Rubisco contents in rice leaves.Plant Cell Environ. 2012 Mar;35(3):546-53. doi: 10.1111/j.1365-3040.2011.02434.x. Epub 2011 Oct 24. Plant Cell Environ. 2012. PMID: 21951138
-
Ribulose 1,5-bisphosphate carboxylase/oxygenase large subunit translation is regulated in a small subunit-independent manner in the expanded leaves of tobacco.Plant Cell Physiol. 2008 Feb;49(2):214-25. doi: 10.1093/pcp/pcm179. Epub 2008 Jan 4. Plant Cell Physiol. 2008. PMID: 18178584
-
Rubisco gene expression in C4 plants.J Exp Bot. 2008;59(7):1625-34. doi: 10.1093/jxb/erm368. Epub 2008 Mar 5. J Exp Bot. 2008. PMID: 18325924 Review.
-
The Hidden Face of Rubisco.Trends Plant Sci. 2018 May;23(5):382-392. doi: 10.1016/j.tplants.2018.02.006. Epub 2018 Mar 7. Trends Plant Sci. 2018. PMID: 29525130 Review.
Cited by
-
Biochemical and Molecular Characterization of Musa sp. Cultured in Temporary Immersion Bioreactor.Plants (Basel). 2023 Nov 4;12(21):3770. doi: 10.3390/plants12213770. Plants (Basel). 2023. PMID: 37960126 Free PMC article.
-
Phytochrome-induced SIG2 expression contributes to photoregulation of phytochrome signalling and photomorphogenesis in Arabidopsis thaliana.J Exp Bot. 2013 Dec;64(18):5457-72. doi: 10.1093/jxb/ert308. Epub 2013 Sep 27. J Exp Bot. 2013. PMID: 24078666 Free PMC article.
-
Cotton bracts are adapted to a microenvironment of concentrated CO2 produced by rapid fruit respiration.Ann Bot. 2013 Jul;112(1):31-40. doi: 10.1093/aob/mct091. Epub 2013 Apr 26. Ann Bot. 2013. PMID: 23625144 Free PMC article.
-
Temperature-induced changes in Arabidopsis Rubisco activity and isoform expression.J Exp Bot. 2023 Jan 11;74(2):651-663. doi: 10.1093/jxb/erac379. J Exp Bot. 2023. PMID: 36124740 Free PMC article.
-
Effects of co-overexpression of the genes of Rubisco and transketolase on photosynthesis in rice.Photosynth Res. 2017 Mar;131(3):281-289. doi: 10.1007/s11120-016-0320-4. Epub 2016 Nov 5. Photosynth Res. 2017. PMID: 27817054
References
-
- Bate NJ, Rothstein SJ, Thompson JE. (1991) Expression of nuclear and chloroplast photosynthetic-specific genes during leaf senescence. J Exp Bot 42: 801–811
-
- Bollenbach TJ, Sharwood RE, Gutierrez R, Lerbs-Mache S, Stern DB. (2009) The RNA-binding proteins CSP41a and CSP41b may regulate transcription and translation of chloroplast-encoded RNAs in Arabidopsis. Plant Mol Biol 69: 541–552 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

