TNF receptor 1 genetic risk mirrors outcome of anti-TNF therapy in multiple sclerosis
- PMID: 22801493
- PMCID: PMC4268493
- DOI: 10.1038/nature11307
TNF receptor 1 genetic risk mirrors outcome of anti-TNF therapy in multiple sclerosis
Abstract
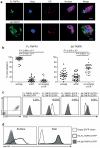
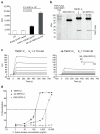
Although there has been much success in identifying genetic variants associated with common diseases using genome-wide association studies (GWAS), it has been difficult to demonstrate which variants are causal and what role they have in disease. Moreover, the modest contribution that these variants make to disease risk has raised questions regarding their medical relevance. Here we have investigated a single nucleotide polymorphism (SNP) in the TNFRSF1A gene, that encodes tumour necrosis factor receptor 1 (TNFR1), which was discovered through GWAS to be associated with multiple sclerosis (MS), but not with other autoimmune conditions such as rheumatoid arthritis, psoriasis and Crohn’s disease. By analysing MS GWAS data in conjunction with the 1000 Genomes Project data we provide genetic evidence that strongly implicates this SNP, rs1800693, as the causal variant in the TNFRSF1A region. We further substantiate this through functional studies showing that the MS risk allele directs expression of a novel, soluble form of TNFR1 that can block TNF. Importantly, TNF-blocking drugs can promote onset or exacerbation of MS, but they have proven highly efficacious in the treatment of autoimmune diseases for which there is no association with rs1800693. This indicates that the clinical experience with these drugs parallels the disease association of rs1800693, and that the MS-associated TNFR1 variant mimics the effect of TNF-blocking drugs. Hence, our study demonstrates that clinical practice can be informed by comparing GWAS across common autoimmune diseases and by investigating the functional consequences of the disease-associated genetic variation.
Figures

Comment in
-
Multiple sclerosis: TNF receptor 1 gene variant could explain failure of TNF-blocking drugs in multiple sclerosis.Nat Rev Neurol. 2012 Sep;8(9):476. doi: 10.1038/nrneurol.2012.149. Epub 2012 Jul 24. Nat Rev Neurol. 2012. PMID: 22825703 No abstract available.
Similar articles
-
Clinical relevance and functional consequences of the TNFRSF1A multiple sclerosis locus.Neurology. 2013 Nov 26;81(22):1891-9. doi: 10.1212/01.wnl.0000436612.66328.8a. Epub 2013 Oct 30. Neurology. 2013. PMID: 24174586 Free PMC article.
-
TNFRSF1A polymorphisms rs1800693 and rs4149584 in patients with multiple sclerosis.Neurology. 2013 May 28;80(22):2010-6. doi: 10.1212/WNL.0b013e318294b2d6. Epub 2013 Apr 26. Neurology. 2013. PMID: 23624563
-
Association of TNFAIP3 and TNFRSF1A variation with multiple sclerosis in a German case-control cohort.Int J Immunogenet. 2015 Apr;42(2):106-10. doi: 10.1111/iji.12183. Epub 2015 Feb 12. Int J Immunogenet. 2015. PMID: 25684197
-
The tumor necrosis factor receptor superfamily member 1B polymorphisms predict response to anti-TNF therapy in patients with autoimmune disease: A meta-analysis.Int Immunopharmacol. 2015 Sep;28(1):146-53. doi: 10.1016/j.intimp.2015.05.049. Epub 2015 Jun 9. Int Immunopharmacol. 2015. PMID: 26071216 Review.
-
The Link Between CD6 and Autoimmunity: Genetic and Cellular Associations.Curr Drug Targets. 2016;17(6):651-65. doi: 10.2174/1389450117666160201105934. Curr Drug Targets. 2016. PMID: 26844569 Review.
Cited by
-
Gene-Based Tests of a Genome-Wide Association Study Dataset Highlight Novel Multiple Sclerosis Risk Genes.Front Neurosci. 2021 May 11;15:614528. doi: 10.3389/fnins.2021.614528. eCollection 2021. Front Neurosci. 2021. PMID: 34045940 Free PMC article.
-
A catalog of GWAS fine-mapping efforts in autoimmune disease.Am J Hum Genet. 2021 Apr 1;108(4):549-563. doi: 10.1016/j.ajhg.2021.03.009. Am J Hum Genet. 2021. PMID: 33798443 Free PMC article. Review.
-
Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci.Nat Genet. 2013 Jul;45(7):730-8. doi: 10.1038/ng.2667. Epub 2013 Jun 9. Nat Genet. 2013. PMID: 23749187 Free PMC article.
-
A "candidate-interactome" aggregate analysis of genome-wide association data in multiple sclerosis.PLoS One. 2013 May 16;8(5):e63300. doi: 10.1371/journal.pone.0063300. Print 2013. PLoS One. 2013. PMID: 23696811 Free PMC article.
-
Inflammatory Cytokines Associated with Multiple Sclerosis Directly Induce Alterations of Neuronal Cytoarchitecture in Human Neurons.J Neuroimmune Pharmacol. 2023 Jun;18(1-2):145-159. doi: 10.1007/s11481-023-10059-w. Epub 2023 Mar 2. J Neuroimmune Pharmacol. 2023. PMID: 36862362 Free PMC article.
References
For ONLINE METHODS
-
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nature Genet. 2007;39:906–913. - PubMed
-
- Chan FK, et al. Fluorescence resonance energy transfer analysis of cell surface receptor interactions and signaling using spectral variants of the green fluorescent protein. Cytometry. 2001;44:361–368. - PubMed
-
- James JR, Oliveira MI, Carmo AM, Iaboni A, Davis SJ. A rigorous experimental framework for detecting protein oligomerization using bioluminescence resonance energy transfer. Nature Methods. 2006;3:1001–1006. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

