Specific interactions between the Candida albicans ABC transporter Cdr1p ectodomain and a D-octapeptide derivative inhibitor
- PMID: 22788839
- PMCID: PMC3418399
- DOI: 10.1111/j.1365-2958.2012.08140.x
Specific interactions between the Candida albicans ABC transporter Cdr1p ectodomain and a D-octapeptide derivative inhibitor
Abstract
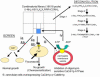
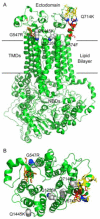
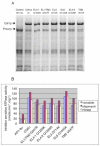
Overexpression of the Candida albicans ATP-binding cassette transporter CaCdr1p causes clinically significant resistance to azole drugs including fluconazole (FLC). Screening of a ~1.89 × 10(6) member D-octapeptide combinatorial library that concentrates library members at the yeast cell surface identified RC21v3, a 4-methoxy-2,3,6-trimethylbenzenesulphonyl derivative of the D-octapeptide D-NH(2) -FFKWQRRR-CONH(2) , as a potent and stereospecific inhibitor of CaCdr1p. RC21v3 chemosensitized Saccharomyces cerevisiae strains overexpressing CaCdr1p but not other fungal ABC transporters, the C. albicans MFS transporter CaMdr1p or the azole target enzyme CaErg11p, to FLC. RC21v3 also chemosensitized clinical C. albicans isolates overexpressing CaCDR1 to FLC, even when CaCDR2 was overexpressed. Specific targeting of CaCdr1p by RC21v3 was confirmed by spontaneous RC21v3 chemosensitization-resistant suppressor mutants of S. cerevisiae expressing CaCdr1p. The suppressor mutations introduced a positive charge beside, or within, extracellular loops 1, 3, 4 and 6 of CaCdr1p or an aromatic residue near the extracytoplasmic end of transmembrane segment 5. The mutations did not affect CaCdr1p localization or CaCdr1p ATPase activity but some increased susceptibility to the CaCdr1p substrates FLC, rhodamine 6G, rhodamine 123 and cycloheximide. The suppressor mutations showed that the drug-like CaCdr1p inhibitors FK506, enniatin, milbemycin α11 and milbemycin β9 have modes of action similar to RC21v3.
© 2012 Blackwell Publishing Ltd.
Figures
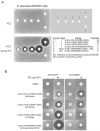


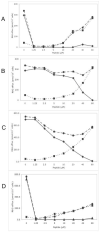
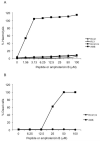
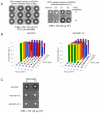
 or absence
or absence  of glucose. The glucose-sensitive efflux remaining after RC21 treatment is shown
of glucose. The glucose-sensitive efflux remaining after RC21 treatment is shown  .
.
Similar articles
-
Inhibitors of the Candida albicans Major Facilitator Superfamily Transporter Mdr1p Responsible for Fluconazole Resistance.PLoS One. 2015 May 7;10(5):e0126350. doi: 10.1371/journal.pone.0126350. eCollection 2015. PLoS One. 2015. PMID: 25951180 Free PMC article.
-
The monoamine oxidase A inhibitor clorgyline is a broad-spectrum inhibitor of fungal ABC and MFS transporter efflux pump activities which reverses the azole resistance of Candida albicans and Candida glabrata clinical isolates.Antimicrob Agents Chemother. 2012 Mar;56(3):1508-15. doi: 10.1128/AAC.05706-11. Epub 2011 Dec 27. Antimicrob Agents Chemother. 2012. PMID: 22203607 Free PMC article.
-
Fungicidal action of geraniol against Candida albicans is potentiated by abrogated CaCdr1p drug efflux and fluconazole synergism.PLoS One. 2018 Aug 29;13(8):e0203079. doi: 10.1371/journal.pone.0203079. eCollection 2018. PLoS One. 2018. PMID: 30157240 Free PMC article.
-
Modulators of the Efflux Pump Cdr1p of Candida albicans: Mechanisms of Action and Chemical Features.Curr Med Chem. 2017;24(30):3242-3253. doi: 10.2174/0929867324666170523102244. Curr Med Chem. 2017. PMID: 28545374 Review.
-
Candida Efflux ATPases and Antiporters in Clinical Drug Resistance.Adv Exp Med Biol. 2016;892:351-376. doi: 10.1007/978-3-319-25304-6_15. Adv Exp Med Biol. 2016. PMID: 26721282 Review.
Cited by
-
Turning Inside Out: Filamentous Fungal Secretion and Its Applications in Biotechnology, Agriculture, and the Clinic.J Fungi (Basel). 2021 Jul 2;7(7):535. doi: 10.3390/jof7070535. J Fungi (Basel). 2021. PMID: 34356914 Free PMC article. Review.
-
Inhibitors of the Candida albicans Major Facilitator Superfamily Transporter Mdr1p Responsible for Fluconazole Resistance.PLoS One. 2015 May 7;10(5):e0126350. doi: 10.1371/journal.pone.0126350. eCollection 2015. PLoS One. 2015. PMID: 25951180 Free PMC article.
-
Targeting efflux pumps to overcome antifungal drug resistance.Future Med Chem. 2016 Aug;8(12):1485-501. doi: 10.4155/fmc-2016-0050. Epub 2016 Jul 27. Future Med Chem. 2016. PMID: 27463566 Free PMC article. Review.
-
Azole Resistance Reduces Susceptibility to the Tetrazole Antifungal VT-1161.Antimicrob Agents Chemother. 2018 Dec 21;63(1):e02114-18. doi: 10.1128/AAC.02114-18. Print 2019 Jan. Antimicrob Agents Chemother. 2018. PMID: 30397057 Free PMC article.
-
Efflux pump proteins in antifungal resistance.Front Pharmacol. 2014 Aug 29;5:202. doi: 10.3389/fphar.2014.00202. eCollection 2014. Front Pharmacol. 2014. PMID: 25221515 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

