RANK (TNFRSF11A) is epigenetically inactivated and induces apoptosis in gliomas
- PMID: 22787434
- PMCID: PMC3394195
- DOI: 10.1596/neo.12360
RANK (TNFRSF11A) is epigenetically inactivated and induces apoptosis in gliomas
Abstract
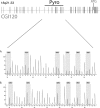
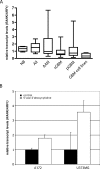
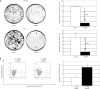
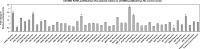
Alterations of DNA methylation play an important role in gliomas. In a genome-wide screen, we identified a CpG-rich fragment within the 5' region of the tumor necrosis factor receptor superfamily, member 11A gene (TNFRSF11A) that showed de novo methylation in gliomas. TNFRSF11A, also known as receptor activator of NF-κB (RANK), activates several signaling pathways, such as NF-κB, JNK, ERK, p38α, and Akt/PKB. Using pyrosequencing, we detected RANK/TNFRSF11A promoter methylation in 8 (57.1%) of 14 diffuse astrocytomas, 17 (77.3%) of 22 anaplastic astrocytomas, 101 (84.2%) of 120 glioblastomas, 6 (100%) of 6 glioma cell lines, and 7 (100%) of 7 glioma stem cell-enriched glioblastoma primary cultures but not in four normal white matter tissue samples. Treatment of glioma cell lines with the demethylating agent 5-aza-2'-deoxycytidine significantly reduced the methylation level and resulted in increased RANK/TNFRSF11A mRNA expression. Overexpression of RANK/TNFRSF11A in glioblastoma cell lines leads to a significant reduction in focus formation and elevated apoptotic activity after flow cytometric analysis. Reporter assay studies of transfected glioma cells supported these results by showing the activation of signaling pathways associated with regulation of apoptosis. We conclude that RANK/TNFRSF11A is a novel and frequent target for de novo methylation in gliomas, which affects apoptotic activity and focus formation thereby contributing to the molecular pathogenesis of gliomas.
Figures

Similar articles
-
Frequent epigenetic inactivation of the chaperone SGNE1/7B2 in human gliomas.Int J Cancer. 2012 Aug 1;131(3):612-22. doi: 10.1002/ijc.26416. Epub 2011 Oct 5. Int J Cancer. 2012. PMID: 21901745
-
Identification of novel human receptor activator of nuclear factor-kB isoforms generated through alternative splicing: implications in breast cancer cell survival and migration.Breast Cancer Res. 2012 Jul 23;14(4):R112. doi: 10.1186/bcr3234. Breast Cancer Res. 2012. PMID: 22824341 Free PMC article.
-
Epigenetic downregulation of mitogen-activated protein kinase phosphatase MKP-2 relieves its growth suppressive activity in glioma cells.Cancer Res. 2010 Feb 15;70(4):1689-99. doi: 10.1158/0008-5472.CAN-09-3218. Epub 2010 Feb 2. Cancer Res. 2010. PMID: 20124482
-
O6-methylguanine DNA methyltransferase gene promoter methylation status in gliomas and its correlation with other molecular alterations: first Indian report with review of challenges for use in customized treatment.Neurosurgery. 2010 Dec;67(6):1681-91. doi: 10.1227/NEU.0b013e3181f743f5. Neurosurgery. 2010. PMID: 21107199 Review.
-
Overview of DNA methylation in adult diffuse gliomas.Brain Tumor Pathol. 2019 Apr;36(2):84-91. doi: 10.1007/s10014-019-00339-w. Epub 2019 Apr 1. Brain Tumor Pathol. 2019. PMID: 30937703 Review.
Cited by
-
Novel Insights reveal Anti-microbial Gene Regulation of Piglet Intestine Immune in response to Clostridium perfringens Infection.Sci Rep. 2019 Feb 13;9(1):1963. doi: 10.1038/s41598-018-37898-5. Sci Rep. 2019. PMID: 30760749 Free PMC article.
-
Identification of a prognostic 5-Gene expression signature for gastric cancer.J Cancer Res Clin Oncol. 2017 Apr;143(4):619-629. doi: 10.1007/s00432-016-2324-z. Epub 2016 Dec 29. J Cancer Res Clin Oncol. 2017. PMID: 28035468
-
Anticancer Mechanism of Curcumin on Human Glioblastoma.Nutrients. 2021 Mar 16;13(3):950. doi: 10.3390/nu13030950. Nutrients. 2021. PMID: 33809462 Free PMC article. Review.
-
Screening and evaluation of the role of immune genes of brain metastasis in lung adenocarcinoma progression based on the TCGA and GEO databases.J Thorac Dis. 2021 Aug;13(8):5016-5034. doi: 10.21037/jtd-21-935. J Thorac Dis. 2021. PMID: 34527340 Free PMC article.
-
MEG-3-mediated Wnt/β-catenin signaling pathway controls the inhibition of tunicamycin-mediated viability in glioblastoma.Oncol Lett. 2018 Sep;16(3):2797-2804. doi: 10.3892/ol.2018.9048. Epub 2018 Jun 28. Oncol Lett. 2018. PMID: 30127865 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
