An essential novel component of the noncanonical mitochondrial outer membrane protein import system of trypanosomatids
- PMID: 22787278
- PMCID: PMC3431924
- DOI: 10.1091/mbc.E12-02-0107
An essential novel component of the noncanonical mitochondrial outer membrane protein import system of trypanosomatids
Abstract
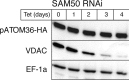
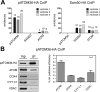
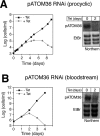
The mitochondrial outer membrane protein Tom40 is the general entry gate for imported proteins in essentially all eukaryotes. Trypanosomatids lack Tom40, however, and use instead a protein termed the archaic translocase of the outer mitochondrial membrane (ATOM). Here we report the discovery of pATOM36, a novel essential component of the trypanosomal outer membrane protein import system that interacts with ATOM. pATOM36 is not related to known Tom proteins from other organisms and mediates the import of matrix proteins. However, there is a group of precursor proteins whose import is independent of pATOM36. Domain-swapping experiments indicate that the N-terminal presequence-containing domain of the substrate proteins at least in part determines the dependence on pATOM36. Secondary structure profiling suggests that pATOM36 is composed largely of α-helices and its assembly into the outer membrane is independent of the sorting and assembly machinery complex. Taken together, these results show that pATOM36 is a novel component associated with the ATOM complex that promotes the import of a subpopulation of proteins into the mitochondrial matrix.
Figures



Similar articles
-
Independent evolution of functionally exchangeable mitochondrial outer membrane import complexes.Elife. 2018 Jun 20;7:e34488. doi: 10.7554/eLife.34488. Elife. 2018. PMID: 29923829 Free PMC article.
-
Outer membrane protein functions as integrator of protein import and DNA inheritance in mitochondria.Proc Natl Acad Sci U S A. 2016 Aug 2;113(31):E4467-75. doi: 10.1073/pnas.1605497113. Epub 2016 Jul 19. Proc Natl Acad Sci U S A. 2016. PMID: 27436903 Free PMC article.
-
Mitochondrial preprotein translocase of trypanosomatids has a bacterial origin.Curr Biol. 2011 Oct 25;21(20):1738-43. doi: 10.1016/j.cub.2011.08.060. Epub 2011 Oct 13. Curr Biol. 2011. PMID: 22000100
-
Tim17 Updates: A Comprehensive Review of an Ancient Mitochondrial Protein Translocator.Biomolecules. 2020 Dec 7;10(12):1643. doi: 10.3390/biom10121643. Biomolecules. 2020. PMID: 33297490 Free PMC article. Review.
-
How does the TOM complex mediate insertion of precursor proteins into the mitochondrial outer membrane?J Cell Biol. 2005 Nov 7;171(3):419-23. doi: 10.1083/jcb.200507147. Epub 2005 Oct 31. J Cell Biol. 2005. PMID: 16260501 Free PMC article. Review.
Cited by
-
Mitochondrial protein import in trypanosomatids: Variations on a theme or fundamentally different?PLoS Pathog. 2018 Nov 29;14(11):e1007351. doi: 10.1371/journal.ppat.1007351. eCollection 2018 Nov. PLoS Pathog. 2018. PMID: 30496284 Free PMC article. Review. No abstract available.
-
Trypanosome alternative oxidase possesses both an N-terminal and internal mitochondrial targeting signal.Eukaryot Cell. 2014 Apr;13(4):539-47. doi: 10.1128/EC.00312-13. Epub 2014 Feb 21. Eukaryot Cell. 2014. PMID: 24562910 Free PMC article.
-
Independent evolution of functionally exchangeable mitochondrial outer membrane import complexes.Elife. 2018 Jun 20;7:e34488. doi: 10.7554/eLife.34488. Elife. 2018. PMID: 29923829 Free PMC article.
-
Is the mitochondrion a promising drug target in trypanosomatids?Mem Inst Oswaldo Cruz. 2022 Feb 21;117:e210379. doi: 10.1590/0074-02760210379. eCollection 2022. Mem Inst Oswaldo Cruz. 2022. PMID: 35195164 Free PMC article.
-
Protein import complexes in the mitochondrial outer membrane of Amoebozoa representatives.BMC Genomics. 2016 Feb 6;17:99. doi: 10.1186/s12864-016-2402-2. BMC Genomics. 2016. PMID: 26852331 Free PMC article.
References
-
- Abe Y, Shodai T, Muto T, Mihara K, Torii H, Nishikawa S-I, Endo T, Kohda D. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell. 2000;100:551–560. - PubMed
-
- Allen R, Egan B, Gabriel K, Beilharz T, Lithgow T. A conserved proline residue is present in the transmembrane-spanning domain of Tom7 and other tail-anchored protein subunits of the TOM translocase. FEBS Lett. 2002;514:347–350. - PubMed
-
- Baker KP, Schatz G. Mitochondrial proteins essential for viability mediate protein import into yeast mitochondria. Nature. 1991;349:205–208. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

