Cysteine peptidases and their inhibitors in Tetranychus urticae: a comparative genomic approach
- PMID: 22784002
- PMCID: PMC3407033
- DOI: 10.1186/1471-2164-13-307
Cysteine peptidases and their inhibitors in Tetranychus urticae: a comparative genomic approach
Abstract
Background: Cysteine peptidases in the two-spotted spider mite Tetranychus urticae are involved in essential physiological processes, including proteolytic digestion. Cystatins and thyropins are inhibitors of cysteine peptidases that modulate their activity, although their function in this species has yet to be investigated. Comparative genomic analyses are powerful tools to obtain advanced knowledge into the presence and evolution of both, peptidases and their inhibitors, and could aid to elucidate issues concerning the function of these proteins.
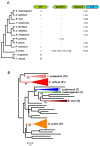
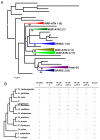
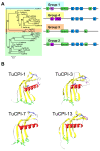
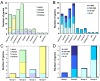
Results: We have performed a genomic comparative analysis of cysteine peptidases and their inhibitors in T. urticae and representative species of different arthropod taxonomic groups. The results indicate: i) clade-specific proliferations are common to C1A papain-like peptidases and for the I25B cystatin family of inhibitors, whereas the C1A inhibitors thyropins are evolutionarily more conserved among arthropod clades; ii) an unprecedented extensive expansion for C13 legumain-like peptidases is found in T. urticae; iii) a sequence-structure analysis of the spider mite cystatins suggests that diversification may be related to an expansion of their inhibitory range; and iv) an in silico transcriptomic analysis shows that most cathepsin B and L cysteine peptidases, legumains and several members of the cystatin family are expressed at a higher rate in T. urticae feeding stages than in embryos.
Conclusion: Comparative genomics has provided valuable insights on the spider mite cysteine peptidases and their inhibitors. Mite-specific proliferations of C1A and C13 peptidase and I25 cystatin families and their over-expression in feeding stages of mites fit with a putative role in mite's feeding and could have a key role in its broad host feeding range.
Figures

Similar articles
-
Digestive proteases in bodies and faeces of the two-spotted spider mite, Tetranychus urticae.J Insect Physiol. 2015 Jul;78:69-77. doi: 10.1016/j.jinsphys.2015.05.002. Epub 2015 May 7. J Insect Physiol. 2015. PMID: 25960286
-
Gene pyramiding of peptidase inhibitors enhances plant resistance to the spider mite Tetranychus urticae.PLoS One. 2012;7(8):e43011. doi: 10.1371/journal.pone.0043011. Epub 2012 Aug 10. PLoS One. 2012. PMID: 22900081 Free PMC article.
-
Characterization of the entire cystatin gene family in barley and their target cathepsin L-like cysteine-proteases, partners in the hordein mobilization during seed germination.Plant Physiol. 2009 Nov;151(3):1531-45. doi: 10.1104/pp.109.146019. Epub 2009 Sep 16. Plant Physiol. 2009. PMID: 19759340 Free PMC article.
-
Cysteine peptidases of mammals: their biological roles and potential effects in the oral cavity and other tissues in health and disease.Crit Rev Oral Biol Med. 2002;13(3):238-75. doi: 10.1177/154411130201300304. Crit Rev Oral Biol Med. 2002. PMID: 12090464 Review.
-
C1A cysteine protease-cystatin interactions in leaf senescence.J Exp Bot. 2014 Jul;65(14):3825-33. doi: 10.1093/jxb/eru043. Epub 2014 Mar 5. J Exp Bot. 2014. PMID: 24600023 Review.
Cited by
-
Meta-transcriptomics indicates biotic cross-tolerance in willow trees cultivated on petroleum hydrocarbon contaminated soil.BMC Plant Biol. 2015 Oct 12;15:246. doi: 10.1186/s12870-015-0636-9. BMC Plant Biol. 2015. PMID: 26459343 Free PMC article.
-
Genomes of trombidid mites reveal novel predicted allergens and laterally transferred genes associated with secondary metabolism.Gigascience. 2018 Dec 1;7(12):giy127. doi: 10.1093/gigascience/giy127. Gigascience. 2018. PMID: 30445460 Free PMC article.
-
Identification of Proteases and Protease Inhibitors in Seeds of the Recalcitrant Forest Tree Species Quercus ilex.Front Plant Sci. 2022 Jun 27;13:907042. doi: 10.3389/fpls.2022.907042. eCollection 2022. Front Plant Sci. 2022. PMID: 35832232 Free PMC article.
-
Phylogenetic relationships, stage-specific expression and localisation of a unique family of inactive cysteine proteases in Sarcoptes scabiei.Parasit Vectors. 2018 May 16;11(1):301. doi: 10.1186/s13071-018-2862-0. Parasit Vectors. 2018. PMID: 29769145 Free PMC article.
-
RNA-Binding Proteins in Trichomonas vaginalis: Atypical Multifunctional Proteins.Biomolecules. 2015 Nov 26;5(4):3354-95. doi: 10.3390/biom5043354. Biomolecules. 2015. PMID: 26703754 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions

