Cohesin regulates tissue-specific expression by stabilizing highly occupied cis-regulatory modules
- PMID: 22780989
- PMCID: PMC3483546
- DOI: 10.1101/gr.136507.111
Cohesin regulates tissue-specific expression by stabilizing highly occupied cis-regulatory modules
Abstract
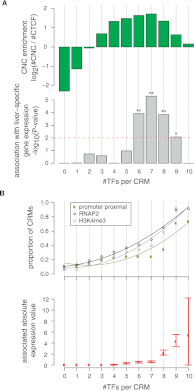
The cohesin protein complex contributes to transcriptional regulation in a CTCF-independent manner by colocalizing with master regulators at tissue-specific loci. The regulation of transcription involves the concerted action of multiple transcription factors (TFs) and cohesin's role in this context of combinatorial TF binding remains unexplored. To investigate cohesin-non-CTCF (CNC) binding events in vivo we mapped cohesin and CTCF, as well as a collection of tissue-specific and ubiquitous transcriptional regulators using ChIP-seq in primary mouse liver. We observe a positive correlation between the number of distinct TFs bound and the presence of CNC sites. In contrast to regions of the genome where cohesin and CTCF colocalize, CNC sites coincide with the binding of master regulators and enhancer-markers and are significantly associated with liver-specific expressed genes. We also show that cohesin presence partially explains the commonly observed discrepancy between TF motif score and ChIP signal. Evidence from these statistical analyses in wild-type cells, and comparisons to maps of TF binding in Rad21-cohesin haploinsufficient mouse liver, suggests that cohesin helps to stabilize large protein-DNA complexes. Finally, we observe that the presence of mirrored CTCF binding events at promoters and their nearby cohesin-bound enhancers is associated with elevated expression levels.
Figures





Similar articles
-
Genome-wide and parental allele-specific analysis of CTCF and cohesin DNA binding in mouse brain reveals a tissue-specific binding pattern and an association with imprinted differentially methylated regions.Genome Res. 2013 Oct;23(10):1624-35. doi: 10.1101/gr.150136.112. Epub 2013 Jun 26. Genome Res. 2013. PMID: 23804403 Free PMC article.
-
Role of CCCTC binding factor (CTCF) and cohesin in the generation of single-cell diversity of protocadherin-α gene expression.Proc Natl Acad Sci U S A. 2012 Jun 5;109(23):9125-30. doi: 10.1073/pnas.1205074109. Epub 2012 May 1. Proc Natl Acad Sci U S A. 2012. PMID: 22550178 Free PMC article.
-
A role for CTCF and cohesin in subtelomere chromatin organization, TERRA transcription, and telomere end protection.EMBO J. 2012 Nov 5;31(21):4165-78. doi: 10.1038/emboj.2012.266. Epub 2012 Sep 25. EMBO J. 2012. PMID: 23010778 Free PMC article.
-
CTCF and cohesin: linking gene regulatory elements with their targets.Cell. 2013 Mar 14;152(6):1285-97. doi: 10.1016/j.cell.2013.02.029. Cell. 2013. PMID: 23498937 Review.
-
Genome-wide studies of CCCTC-binding factor (CTCF) and cohesin provide insight into chromatin structure and regulation.J Biol Chem. 2012 Sep 7;287(37):30906-13. doi: 10.1074/jbc.R111.324962. Epub 2012 Sep 5. J Biol Chem. 2012. PMID: 22952237 Free PMC article. Review.
Cited by
-
GENE REGULATION. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock.Science. 2015 Jun 26;348(6242):1488-92. doi: 10.1126/science.aab3021. Epub 2015 Jun 4. Science. 2015. PMID: 26044300 Free PMC article.
-
Topoisomerase II beta interacts with cohesin and CTCF at topological domain borders.Genome Biol. 2016 Aug 31;17(1):182. doi: 10.1186/s13059-016-1043-8. Genome Biol. 2016. PMID: 27582050 Free PMC article.
-
Insulator function and topological domain border strength scale with architectural protein occupancy.Genome Biol. 2014 Jun 30;15(6):R82. doi: 10.1186/gb-2014-15-5-r82. Genome Biol. 2014. PMID: 24981874 Free PMC article.
-
The 3D genome in transcriptional regulation and pluripotency.Cell Stem Cell. 2014 Jun 5;14(6):762-75. doi: 10.1016/j.stem.2014.05.017. Cell Stem Cell. 2014. PMID: 24905166 Free PMC article. Review.
-
Separase prevents genomic instability by controlling replication fork speed.Nucleic Acids Res. 2018 Jan 9;46(1):267-278. doi: 10.1093/nar/gkx1172. Nucleic Acids Res. 2018. PMID: 29165708 Free PMC article.
References
-
- Adcock IM, Caramori G 2001. Cross-talk between pro-inflammatory transcription factors and glucocorticoids. Immunol Cell Biol 79: 376–384 - PubMed
-
- Bailey TL, Elkan C 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
