Regulation of DCIS to invasive breast cancer progression by Singleminded-2s (SIM2s)
- PMID: 22777354
- PMCID: PMC4469191
- DOI: 10.1038/onc.2012.286
Regulation of DCIS to invasive breast cancer progression by Singleminded-2s (SIM2s)
Abstract
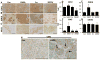
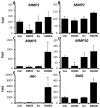
Singleminded-2s (SIM2s) is a member of the bHLH/PAS family of transcription factors and a key regulator of mammary epithelial cell differentiation. SIM2s is highly expressed in mammary epithelial cells and downregulated in human breast cancer. Loss of Sim2s causes aberrant mouse mammary ductal development, with features suggestive of malignant transformation, whereas overexpression of SIM2s promotes precocious alveolar differentiation in nulliparous mouse mammary glands, suggesting that SIM2s is required for establishing and enhancing mammary gland differentiation. To test the hypothesis that SIM2s regulates tumor cell differentiation, we analyzed SIM2s expression in human primary breast ductal carcinoma in situ (DCIS) samples and found that SIM2s is lost with progression from DCIS to invasive ductal cancer (IDC). Using a MCF10DCIS.COM progression model, we have shown that SIM2s expression is decreased in MCF10DCIS.COM cells compared with MCF10A cells, and reestablishment of SIM2s in MCF10DCIS.COM cells significantly inhibits growth and invasion both in vitro and in vivo. Analysis of SIM2s-MCF10DCIS.com tumors showed that SIM2s promoted a more differentiated tumor phenotype including the expression of a broad range of luminal markers (CSN2 (β-casein), CDH1 (E-cadherin), and KER18 (keratin-18)) and suppressed genes associated with stem cell maintenance and a basal phenotype (SMO (smoothened), p63, SLUG (snail-2), KER14 (keratin-14) and VIM (vimentin)). Furthermore, loss of SIM2s expression in MCF10DCIS.COM xenografts resulted in a more invasive phenotype and increased lung metastasis likely due to an increase in Hedgehog signaling and matrix metalloproteinase expression. Together, these exciting new data support a role for SIM2s in promoting human breast tumor differentiation and maintaining epithelial integrity.
Conflict of interest statement
The authors have nothing to disclose.
Figures

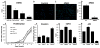



Similar articles
-
Loss of singleminded-2s in the mouse mammary gland induces an epithelial-mesenchymal transition associated with up-regulation of slug and matrix metalloprotease 2.Mol Cell Biol. 2008 Mar;28(6):1936-46. doi: 10.1128/MCB.01701-07. Epub 2007 Dec 26. Mol Cell Biol. 2008. PMID: 18160708 Free PMC article.
-
The bHLH/PAS transcription factor singleminded 2s promotes mammary gland lactogenic differentiation.Development. 2010 Mar;137(6):945-52. doi: 10.1242/dev.041657. Epub 2010 Feb 11. Development. 2010. PMID: 20150276 Free PMC article.
-
Singleminded-2s (Sim2s) promotes delayed involution of the mouse mammary gland through suppression of Stat3 and NFκB.Mol Endocrinol. 2011 Apr;25(4):635-44. doi: 10.1210/me.2010-0423. Epub 2011 Feb 3. Mol Endocrinol. 2011. PMID: 21292822 Free PMC article.
-
Mammary stem cells and breast cancer--role of Notch signalling.Stem Cell Rev. 2007 Jun;3(2):169-75. doi: 10.1007/s12015-007-0023-5. Stem Cell Rev. 2007. PMID: 17873349 Review.
-
Intra-mammary ductal transplantation: a tool to study premalignant progression.J Mammary Gland Biol Neoplasia. 2012 Jun;17(2):131-3. doi: 10.1007/s10911-012-9259-z. Epub 2012 Jun 12. J Mammary Gland Biol Neoplasia. 2012. PMID: 22688217 Review.
Cited by
-
Systems biology comprehensive analysis on breast cancer for identification of key gene modules and genes associated with TNM-based clinical stages.Sci Rep. 2020 Jul 2;10(1):10816. doi: 10.1038/s41598-020-67643-w. Sci Rep. 2020. PMID: 32616754 Free PMC article.
-
Loss of STING impairs lactogenic differentiation.Development. 2024 Oct 1;151(19):dev202998. doi: 10.1242/dev.202998. Epub 2024 Oct 14. Development. 2024. PMID: 39399905 Free PMC article.
-
CD24-associated ceRNA network reveals prognostic biomarkers in breast carcinoma.Sci Rep. 2023 Mar 7;13(1):3826. doi: 10.1038/s41598-022-25072-x. Sci Rep. 2023. PMID: 36882451 Free PMC article.
-
Epiregulin contributes to breast tumorigenesis through regulating matrix metalloproteinase 1 and promoting cell survival.Mol Cancer. 2015 Jul 29;14:138. doi: 10.1186/s12943-015-0408-z. Mol Cancer. 2015. PMID: 26215578 Free PMC article.
-
A nation-wide multicenter retrospective study of the epidemiological, pathological and clinical characteristics of breast cancer in situ in Chinese women in 1999 - 2008.PLoS One. 2013 Nov 20;8(11):e81055. doi: 10.1371/journal.pone.0081055. eCollection 2013. PLoS One. 2013. PMID: 24278375 Free PMC article.
References
-
- Burstein HJ, Polyak K, Wong JS, Lester SC, Kaelin CM. Ductal carcinoma in situ of the breast. N Engl J Med. 2004;350(14):1430–41. Epub 2004/04/09. - PubMed
-
- Cody HS., 3rd Sentinel lymph node biopsy for DCIS: are we approaching consensus? Ann Surg Oncol. 2007;14(8):2179–81. Epub 2007/02/01. - PubMed
-
- Maffuz A, Barroso-Bravo S, Najera I, Zarco G, Alvarado-Cabrero I, Rodriguez-Cuevas SA. Tumor size as predictor of microinvasion, invasion, and axillary metastasis in ductal carcinoma in situ. J Exp Clin Cancer Res. 2006;25(2):223–7. Epub 2006/08/22. - PubMed
-
- Yao J, Weremowicz S, Feng B, Gentleman RC, Marks JR, Gelman R, et al. Combined cDNA array comparative genomic hybridization and serial analysis of gene expression analysis of breast tumor progression. Cancer Res. 2006;66(8):4065–78. Epub 2006/04/19. - PubMed
-
- Chin K, de Solorzano CO, Knowles D, Jones A, Chou W, Rodriguez EG, et al. In situ analyses of genome instability in breast cancer. Nat Genet. 2004;36(9):984–8. Epub 2004/08/10. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

