A map of the cis-regulatory sequences in the mouse genome
- PMID: 22763441
- PMCID: PMC4041622
- DOI: 10.1038/nature11243
A map of the cis-regulatory sequences in the mouse genome
Abstract
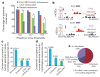
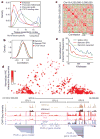
The laboratory mouse is the most widely used mammalian model organism in biomedical research. The 2.6 × 10(9) bases of the mouse genome possess a high degree of conservation with the human genome, so a thorough annotation of the mouse genome will be of significant value to understanding the function of the human genome. So far, most of the functional sequences in the mouse genome have yet to be found, and the cis-regulatory sequences in particular are still poorly annotated. Comparative genomics has been a powerful tool for the discovery of these sequences, but on its own it cannot resolve their temporal and spatial functions. Recently, ChIP-Seq has been developed to identify cis-regulatory elements in the genomes of several organisms including humans, Drosophila melanogaster and Caenorhabditis elegans. Here we apply the same experimental approach to a diverse set of 19 tissues and cell types in the mouse to produce a map of nearly 300,000 murine cis-regulatory sequences. The annotated sequences add up to 11% of the mouse genome, and include more than 70% of conserved non-coding sequences. We define tissue-specific enhancers and identify potential transcription factors regulating gene expression in each tissue or cell type. Finally, we show that much of the mouse genome is organized into domains of coordinately regulated enhancers and promoters. Our results provide a resource for the annotation of functional elements in the mammalian genome and for the study of mechanisms regulating tissue-specific gene expression.
Conflict of interest statement
The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at
Figures


Similar articles
-
A comparative encyclopedia of DNA elements in the mouse genome.Nature. 2014 Nov 20;515(7527):355-64. doi: 10.1038/nature13992. Nature. 2014. PMID: 25409824 Free PMC article.
-
A map of cis-regulatory elements and 3D genome structures in zebrafish.Nature. 2020 Dec;588(7837):337-343. doi: 10.1038/s41586-020-2962-9. Epub 2020 Nov 25. Nature. 2020. PMID: 33239788 Free PMC article.
-
A cis-regulatory map of the Drosophila genome.Nature. 2011 Mar 24;471(7339):527-31. doi: 10.1038/nature09990. Nature. 2011. PMID: 21430782 Free PMC article.
-
Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence.Nat Rev Genet. 2011 Dec 6;13(1):59-69. doi: 10.1038/nrg3095. Nat Rev Genet. 2011. PMID: 22143240 Review.
-
Beyond the ENCODE project: using genomics and epigenomics strategies to study enhancer evolution.Philos Trans R Soc Lond B Biol Sci. 2013 Nov 11;368(1632):20130022. doi: 10.1098/rstb.2013.0022. Print 2013 Dec 19. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 24218635 Free PMC article. Review.
Cited by
-
Microcephaly gene links trithorax and REST/NRSF to control neural stem cell proliferation and differentiation.Cell. 2012 Nov 21;151(5):1097-112. doi: 10.1016/j.cell.2012.10.043. Cell. 2012. PMID: 23178126 Free PMC article.
-
Epigenetic layers and players underlying neurodevelopment.Trends Neurosci. 2013 Aug;36(8):460-70. doi: 10.1016/j.tins.2013.05.001. Epub 2013 May 31. Trends Neurosci. 2013. PMID: 23731492 Free PMC article. Review.
-
Chicken chromatin accessibility atlas accelerates epigenetic annotation of birds and gene fine-mapping associated with growth traits.Zool Res. 2023 Jan 18;44(1):53-62. doi: 10.24272/j.issn.2095-8137.2022.228. Zool Res. 2023. PMID: 36317479 Free PMC article.
-
PETModule: a motif module based approach for enhancer target gene prediction.Sci Rep. 2016 Jul 20;6:30043. doi: 10.1038/srep30043. Sci Rep. 2016. PMID: 27436110 Free PMC article.
-
The contribution of cohesin-SA1 to gene expression and chromatin architecture in two murine tissues.Nucleic Acids Res. 2015 Mar 31;43(6):3056-67. doi: 10.1093/nar/gkv144. Epub 2015 Mar 3. Nucleic Acids Res. 2015. PMID: 25735743 Free PMC article.
References
-
- Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

