Cooperativity among short amyloid stretches in long amyloidogenic sequences
- PMID: 22761773
- PMCID: PMC3382238
- DOI: 10.1371/journal.pone.0039369
Cooperativity among short amyloid stretches in long amyloidogenic sequences
Abstract
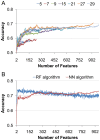
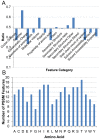
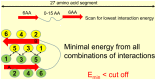
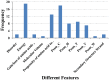
Amyloid fibrillar aggregates of polypeptides are associated with many neurodegenerative diseases. Short peptide segments in protein sequences may trigger aggregation. Identifying these stretches and examining their behavior in longer protein segments is critical for understanding these diseases and obtaining potential therapies. In this study, we combined machine learning and structure-based energy evaluation to examine and predict amyloidogenic segments. Our feature selection method discovered that windows consisting of long amino acid segments of ~30 residues, instead of the commonly used short hexapeptides, provided the highest accuracy. Weighted contributions of an amino acid at each position in a 27 residue window revealed three cooperative regions of short stretch, resemble the β-strand-turn-β-strand motif in A-βpeptide amyloid and β-solenoid structure of HET-s(218-289) prion (C). Using an in-house energy evaluation algorithm, the interaction energy between two short stretches in long segment is computed and incorporated as an additional feature. The algorithm successfully predicted and classified amyloid segments with an overall accuracy of 75%. Our study revealed that genome-wide amyloid segments are not only dependent on short high propensity stretches, but also on nearby residues.
Conflict of interest statement
Figures

Similar articles
-
FISH Amyloid - a new method for finding amyloidogenic segments in proteins based on site specific co-occurrence of aminoacids.BMC Bioinformatics. 2014 Feb 24;15:54. doi: 10.1186/1471-2105-15-54. BMC Bioinformatics. 2014. PMID: 24564523 Free PMC article.
-
A molecular dynamics approach to the structural characterization of amyloid aggregation.J Mol Biol. 2006 Apr 7;357(4):1306-21. doi: 10.1016/j.jmb.2006.01.009. Epub 2006 Jan 26. J Mol Biol. 2006. PMID: 16483608
-
Exploiting heterogeneous features to improve in silico prediction of peptide status - amyloidogenic or non-amyloidogenic.BMC Bioinformatics. 2011;12 Suppl 13(Suppl 13):S21. doi: 10.1186/1471-2105-12-S13-S21. Epub 2011 Nov 30. BMC Bioinformatics. 2011. PMID: 22373069 Free PMC article.
-
Molecular structures of amyloid and prion fibrils: consensus versus controversy.Acc Chem Res. 2013 Jul 16;46(7):1487-96. doi: 10.1021/ar300282r. Epub 2013 Jan 7. Acc Chem Res. 2013. PMID: 23294335 Free PMC article. Review.
-
Predicting the aggregation propensity of prion sequences.Virus Res. 2015 Sep 2;207:127-35. doi: 10.1016/j.virusres.2015.03.001. Epub 2015 Mar 6. Virus Res. 2015. PMID: 25747492 Review.
Cited by
-
Critical Nucleus Structure and Aggregation Mechanism of the C-terminal Fragment of Copper-Zinc Superoxide Dismutase Protein.ACS Chem Neurosci. 2016 Mar 16;7(3):286-96. doi: 10.1021/acschemneuro.5b00242. Epub 2016 Feb 10. ACS Chem Neurosci. 2016. PMID: 26815332 Free PMC article.
-
Self-aggregation and coaggregation of the p53 core fragment with its aggregation gatekeeper variant.Phys Chem Chem Phys. 2016 Mar 21;18(11):8098-107. doi: 10.1039/c5cp06538k. Phys Chem Chem Phys. 2016. PMID: 26923710 Free PMC article.
-
Structural Insight into Tau Protein's Paradox of Intrinsically Disordered Behavior, Self-Acetylation Activity, and Aggregation.J Phys Chem Lett. 2014 Sep 4;5(17):3026-3031. doi: 10.1021/jz501457f. Epub 2014 Aug 19. J Phys Chem Lett. 2014. PMID: 25206938 Free PMC article.
-
Insights into Network of Hot Spots of Aggregation in Nucleophosmin 1.Int J Mol Sci. 2022 Nov 25;23(23):14704. doi: 10.3390/ijms232314704. Int J Mol Sci. 2022. PMID: 36499032 Free PMC article.
-
Prediction of gene phenotypes based on GO and KEGG pathway enrichment scores.Biomed Res Int. 2013;2013:870795. doi: 10.1155/2013/870795. Epub 2013 Nov 7. Biomed Res Int. 2013. PMID: 24312912 Free PMC article.
References
-
- Trojanowski JQ, Mattson MP. Overview of protein aggregation in single, double, and triple neurodegenerative brain amyloidoses. Neuromolecular Med. 2003;4:1–6. - PubMed
-
- Dobson CM. Getting out of shape. Nature. 2002;418:729–730. - PubMed
-
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

