Gene expression profiles in Parkinson disease prefrontal cortex implicate FOXO1 and genes under its transcriptional regulation
- PMID: 22761592
- PMCID: PMC3386245
- DOI: 10.1371/journal.pgen.1002794
Gene expression profiles in Parkinson disease prefrontal cortex implicate FOXO1 and genes under its transcriptional regulation
Abstract
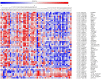
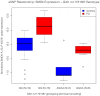
Parkinson disease (PD) is a complex neurodegenerative disorder with largely unknown genetic mechanisms. While the degeneration of dopaminergic neurons in PD mainly takes place in the substantia nigra pars compacta (SN) region, other brain areas, including the prefrontal cortex, develop Lewy bodies, the neuropathological hallmark of PD. We generated and analyzed expression data from the prefrontal cortex Brodmann Area 9 (BA9) of 27 PD and 26 control samples using the 44K One-Color Agilent 60-mer Whole Human Genome Microarray. All samples were male, without significant Alzheimer disease pathology and with extensive pathological annotation available. 507 of the 39,122 analyzed expression probes were different between PD and control samples at false discovery rate (FDR) of 5%. One of the genes with significantly increased expression in PD was the forkhead box O1 (FOXO1) transcription factor. Notably, genes carrying the FoxO1 binding site were significantly enriched in the FDR-significant group of genes (177 genes covered by 189 probes), suggesting a role for FoxO1 upstream of the observed expression changes. Single-nucleotide polymorphisms (SNPs) selected from a recent meta-analysis of PD genome-wide association studies (GWAS) were successfully genotyped in 50 out of the 53 microarray brains, allowing a targeted expression-SNP (eSNP) analysis for 52 SNPs associated with PD affection at genome-wide significance and the 189 probes from FoxO1 regulated genes. A significant association was observed between a SNP in the cyclin G associated kinase (GAK) gene and a probe in the spermine oxidase (SMOX) gene. Further examination of the FOXO1 region in a meta-analysis of six available GWAS showed two SNPs significantly associated with age at onset of PD. These results implicate FOXO1 as a PD-relevant gene and warrant further functional analyses of its transcriptional regulatory mechanisms.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Cyclin-G-associated kinase modifies α-synuclein expression levels and toxicity in Parkinson's disease: results from the GenePD Study.Hum Mol Genet. 2011 Apr 15;20(8):1478-87. doi: 10.1093/hmg/ddr026. Epub 2011 Jan 21. Hum Mol Genet. 2011. PMID: 21258085 Free PMC article.
-
Integrative analyses of proteomics and RNA transcriptomics implicate mitochondrial processes, protein folding pathways and GWAS loci in Parkinson disease.BMC Med Genomics. 2016 Jan 21;9:5. doi: 10.1186/s12920-016-0164-y. BMC Med Genomics. 2016. PMID: 26793951 Free PMC article.
-
Genome-wide analysis of FoxO1 binding in hepatic chromatin: potential involvement of FoxO1 in linking retinoid signaling to hepatic gluconeogenesis.Nucleic Acids Res. 2012 Dec;40(22):11499-509. doi: 10.1093/nar/gks932. Epub 2012 Oct 12. Nucleic Acids Res. 2012. PMID: 23066095 Free PMC article.
-
Quantitative assessment of the association between GAK rs1564282 C/T polymorphism and the risk of Parkinson's disease.J Clin Neurosci. 2015 Jul;22(7):1077-80. doi: 10.1016/j.jocn.2014.12.014. Epub 2015 May 11. J Clin Neurosci. 2015. PMID: 25975492 Review.
-
What genetics tells us about the causes and mechanisms of Parkinson's disease.Physiol Rev. 2011 Oct;91(4):1161-218. doi: 10.1152/physrev.00022.2010. Physiol Rev. 2011. PMID: 22013209 Review.
Cited by
-
Bone marrow-derived mesenchymal stem cells increase dopamine synthesis in the injured striatum.Neural Regen Res. 2012 Dec 5;7(34):2653-62. doi: 10.3969/j.issn.1673-5374.2012.34.002. Neural Regen Res. 2012. PMID: 25337111 Free PMC article.
-
Co-Expression Network Analysis Identifies Molecular Determinants of Loneliness Associated with Neuropsychiatric and Neurodegenerative Diseases.Int J Mol Sci. 2023 Mar 21;24(6):5909. doi: 10.3390/ijms24065909. Int J Mol Sci. 2023. PMID: 36982982 Free PMC article.
-
Convergent Genetic and Expression Datasets Highlight TREM2 in Parkinson's Disease Susceptibility.Mol Neurobiol. 2016 Sep;53(7):4931-8. doi: 10.1007/s12035-015-9416-7. Epub 2015 Sep 14. Mol Neurobiol. 2016. PMID: 26365049
-
Differential Analysis of A-to-I mRNA Edited Sites in Parkinson's Disease.Genes (Basel). 2021 Dec 22;13(1):14. doi: 10.3390/genes13010014. Genes (Basel). 2021. PMID: 35052353 Free PMC article.
-
Sociogenomics of self vs. non-self cooperation during development of Dictyostelium discoideum.BMC Genomics. 2014 Jul 21;15(1):616. doi: 10.1186/1471-2164-15-616. BMC Genomics. 2014. PMID: 25048306 Free PMC article.
References
-
- Lees AJ, Hardy J, Revesz T. Parkinson's disease. Lancet. 2009;373:2055–2066. - PubMed
-
- Ferrer I, Martinez A, Blanco R, Dalfo E, Carmona M. Neuropathology of sporadic Parkinson disease before the appearance of parkinsonism: preclinical Parkinson disease. J Neural Transm. 2011;118:821–839. - PubMed
-
- Sutherland GT, Matigian NA, Chalk AM, Anderson MJ, Silburn PA, et al. A cross-study transcriptional analysis of Parkinson's disease. PLoS ONE. 2009;4:e4955. doi: 10.1371/journal.pone.0004955. - DOI - PMC - PubMed
-
- Edwards YJ, Beecham GW, Scott WK, Khuri S, Bademci G, et al. Identifying consensus disease pathways in Parkinson's disease using an integrative systems biology approach. PLoS ONE. 2011;6:e16917. doi: 10.1371/journal.pone.0016917. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R56 NS037167/NS/NINDS NIH HHS/United States
- R24 MH 068855/MH/NIMH NIH HHS/United States
- R01 CA141668/CA/NCI NIH HHS/United States
- NS036711/NS/NINDS NIH HHS/United States
- P50 NS039764/NS/NINDS NIH HHS/United States
- NS39764/NS/NINDS NIH HHS/United States
- P30 AG19610/AG/NIA NIH HHS/United States
- P30 AG019610/AG/NIA NIH HHS/United States
- R24 MH068855/MH/NIMH NIH HHS/United States
- R01 NS036711/NS/NINDS NIH HHS/United States
- P50 NS071674/NS/NINDS NIH HHS/United States
- R01 NS076843/NS/NINDS NIH HHS/United States
- NS037167/NS/NINDS NIH HHS/United States
- CA141668/CA/NCI NIH HHS/United States
- R01 NS037167/NS/NINDS NIH HHS/United States
- HHSN268200782096C/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

