Endo-β-N-acetylglucosaminidases from infant gut-associated bifidobacteria release complex N-glycans from human milk glycoproteins
- PMID: 22745059
- PMCID: PMC3434770
- DOI: 10.1074/mcp.M112.018119
Endo-β-N-acetylglucosaminidases from infant gut-associated bifidobacteria release complex N-glycans from human milk glycoproteins
Abstract
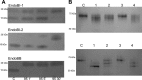
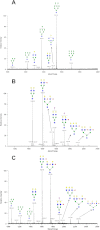
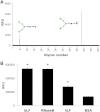
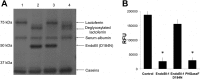
Breastfeeding is one of the main factors guiding the composition of the infant gut microbiota in the first months of life. This process is shaped in part by the high amounts of human milk oligosaccharides that serve as a carbon source for saccharolytic bacteria such as Bifidobacterium species. Infant-borne bifidobacteria have developed various molecular strategies for utilizing these oligosaccharides as a carbon source. We hypothesized that these species also interact with N-glycans found in host glycoproteins that are structurally similar to free oligosaccharides in human milk. Endo-β-N-acetylglucosaminidases were identified in certain isolates of Bifidobacterium longum subsp. longum, B. longum subsp. infantis, and Bifidobacterium breve, and their presence correlated with the ability of these strains to deglycosylate glycoproteins. An endoglycosidase from B. infantis ATCC 15697, EndoBI-1, was active toward all major types of N-linked glycans found in glycosylated proteins. Its activity was not affected by core fucosylation or extensive fucosylation, antenna number, or sialylation, releasing several N-glycans from human lactoferrin and immunoglobulins A and G. Extensive N-deglycosylation of whole breast milk was also observed after coincubation with this enzyme. Mutation of the active site of EndoBI-1 did not abolish binding to N-glycosylated proteins, and this mutant specifically recognized Man(3)GlcNAc(2)(α1-6Fuc), the core structure of human N-glycans. EndoBI-1 is constitutively expressed in B. infantis, and incubation of the bacterium with human or bovine lactoferrin led to the induction of genes associated to import and consumption of human milk oligosaccharides, suggesting linked regulatory mechanisms among these glycans. This work reveals an unprecedented interaction of bifidobacteria with host N-glycans and describes a novel endoglycosidase with broad specificity on diverse N-glycan types, potentially a useful tool for glycoproteomics studies.
Figures

Similar articles
-
Potential Applications of Endo-β-N-Acetylglucosaminidases From Bifidobacterium longum Subspecies infantis in Designing Value-Added, Next-Generation Infant Formulas.Front Nutr. 2021 Apr 9;8:646275. doi: 10.3389/fnut.2021.646275. eCollection 2021. Front Nutr. 2021. PMID: 33898500 Free PMC article. Review.
-
Oligosaccharides Released from Milk Glycoproteins Are Selective Growth Substrates for Infant-Associated Bifidobacteria.Appl Environ Microbiol. 2016 May 31;82(12):3622-3630. doi: 10.1128/AEM.00547-16. Print 2016 Jun 15. Appl Environ Microbiol. 2016. PMID: 27084007 Free PMC article.
-
Kinetic characterization of a novel endo-β-N-acetylglucosaminidase on concentrated bovine colostrum whey to release bioactive glycans.Enzyme Microb Technol. 2015 Sep;77:46-53. doi: 10.1016/j.enzmictec.2015.05.007. Epub 2015 Jun 3. Enzyme Microb Technol. 2015. PMID: 26138399 Free PMC article.
-
Release of bifidogenic N-glycans from native bovine colostrum proteins by an endo-β-N-acetylglucosaminidase.Enzyme Microb Technol. 2023 Jan;162:110138. doi: 10.1016/j.enzmictec.2022.110138. Epub 2022 Oct 4. Enzyme Microb Technol. 2023. PMID: 36252443
-
Bifidobacterium longum subspecies infantis: champion colonizer of the infant gut.Pediatr Res. 2015 Jan;77(1-2):229-35. doi: 10.1038/pr.2014.156. Epub 2014 Oct 10. Pediatr Res. 2015. PMID: 25303277 Free PMC article. Review.
Cited by
-
Utilization of Host-Derived Glycans by Intestinal Lactobacillus and Bifidobacterium Species.Front Microbiol. 2018 Aug 17;9:1917. doi: 10.3389/fmicb.2018.01917. eCollection 2018. Front Microbiol. 2018. PMID: 30177920 Free PMC article. Review.
-
Quantitative Longitudinal Inventory of the N-Glycoproteome of Human Milk from a Single Donor Reveals the Highly Variable Repertoire and Dynamic Site-Specific Changes.J Proteome Res. 2020 May 1;19(5):1941-1952. doi: 10.1021/acs.jproteome.9b00753. Epub 2020 Mar 27. J Proteome Res. 2020. PMID: 32125861 Free PMC article.
-
The impact of the milk glycobiome on the neonate gut microbiota.Annu Rev Anim Biosci. 2015;3:419-45. doi: 10.1146/annurev-animal-022114-111112. Epub 2014 Nov 5. Annu Rev Anim Biosci. 2015. PMID: 25387230 Free PMC article. Review.
-
A novel gene cluster allows preferential utilization of fucosylated milk oligosaccharides in Bifidobacterium longum subsp. longum SC596.Sci Rep. 2016 Oct 19;6:35045. doi: 10.1038/srep35045. Sci Rep. 2016. PMID: 27756904 Free PMC article.
-
Potential Applications of Endo-β-N-Acetylglucosaminidases From Bifidobacterium longum Subspecies infantis in Designing Value-Added, Next-Generation Infant Formulas.Front Nutr. 2021 Apr 9;8:646275. doi: 10.3389/fnut.2021.646275. eCollection 2021. Front Nutr. 2021. PMID: 33898500 Free PMC article. Review.
References
-
- Neville M. C. (2009) Classic studies of mammary development and milk secretion: 1945–1980. J Mammary Gland Biol. Neoplasia 14, 193–197 - PubMed
-
- German J. B., Dillard C. J., Ward R. E. (2002) Bioactive components in milk. Curr. Opin. Clin. Nutr. Metab. Care 5, 653–658 - PubMed
-
- Adlerberth I., Wold A. E. (2009) Establishment of the gut microbiota in Western infants. Acta Paediatr. 98, 229–238 - PubMed
-
- Yatsunenko T., Rey F. E., Manary M. J., Trehan I., Dominguez-Bello M. G., Contreras M., Magris M., Hidalgo G., Baldassano R. N., Anokhin A. P., Heath A. C., Warner B., Reeder J., Kuczynski J., Caporaso J. G., Lozupone C. A., Lauber C., Clemente J. C., Knights D., Knight R., Gordon J. I. (2012) Human gut microbiome viewed across age and geography. Nature 486, 222–227 - PMC - PubMed
-
- Le Huërou-Luron I., Blat S., Boudry G. (2010) Breast- v. formula-feeding: impacts on the digestive tract and immediate and long-term health effects. Nutr. Res. Rev. 23, 23–36 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

