Modular control of multiple pathways using engineered orthogonal T7 polymerases
- PMID: 22743271
- PMCID: PMC3458549
- DOI: 10.1093/nar/gks597
Modular control of multiple pathways using engineered orthogonal T7 polymerases
Abstract
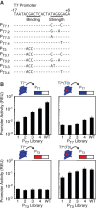
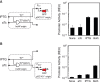
Synthetic genetic sensors and circuits enable programmable control over the timing and conditions of gene expression. They are being increasingly incorporated into the control of complex, multigene pathways and cellular functions. Here, we propose a design strategy to genetically separate the sensing/circuitry functions from the pathway to be controlled. This separation is achieved by having the output of the circuit drive the expression of a polymerase, which then activates the pathway from polymerase-specific promoters. The sensors, circuits and polymerase are encoded together on a 'controller' plasmid. Variants of T7 RNA polymerase that reduce toxicity were constructed and used as scaffolds for the construction of four orthogonal polymerases identified via part mining that bind to unique promoter sequences. This set is highly orthogonal and induces cognate promoters by 8- to 75-fold more than off-target promoters. These orthogonal polymerases enable four independent channels linking the outputs of circuits to the control of different cellular functions. As a demonstration, we constructed a controller plasmid that integrates two inducible systems, implements an AND logic operation and toggles between metabolic pathways that change Escherichia coli green (deoxychromoviridans) and red (lycopene). The advantages of this organization are that (i) the regulation of the pathway can be changed simply by introducing a different controller plasmid, (ii) transcription is orthogonal to host machinery and (iii) the pathway genes are not transcribed in the absence of a controller and are thus more easily carried without invoking evolutionary pressure.
Figures



Similar articles
-
Construction of synthetic T7 RNA polymerase expression systems.Methods. 2018 Jul 1;143:110-120. doi: 10.1016/j.ymeth.2018.02.022. Epub 2018 Mar 5. Methods. 2018. PMID: 29518499
-
A method for cost-effective and rapid characterization of engineered T7-based transcription factors by cell-free protein synthesis reveals insights into the regulation of T7 RNA polymerase-driven expression.Arch Biochem Biophys. 2019 Oct 15;674:108045. doi: 10.1016/j.abb.2019.07.010. Epub 2019 Jul 19. Arch Biochem Biophys. 2019. PMID: 31326518
-
Environmental signal integration by a modular AND gate.Mol Syst Biol. 2007;3:133. doi: 10.1038/msb4100173. Epub 2007 Aug 14. Mol Syst Biol. 2007. PMID: 17700541 Free PMC article.
-
Structure and function in promoter escape by T7 RNA polymerase.Prog Nucleic Acid Res Mol Biol. 2005;80:323-47. doi: 10.1016/S0079-6603(05)80008-X. Prog Nucleic Acid Res Mol Biol. 2005. PMID: 16164978 Review. No abstract available.
-
Construction and Application of Orthogonal T7 Expression System in Eukaryote: An Overview.Adv Biol (Weinh). 2023 Feb;7(2):e2200218. doi: 10.1002/adbi.202200218. Epub 2022 Dec 4. Adv Biol (Weinh). 2023. PMID: 36464626 Review.
Cited by
-
Gas Fermentation-A Flexible Platform for Commercial Scale Production of Low-Carbon-Fuels and Chemicals from Waste and Renewable Feedstocks.Front Microbiol. 2016 May 11;7:694. doi: 10.3389/fmicb.2016.00694. eCollection 2016. Front Microbiol. 2016. PMID: 27242719 Free PMC article. Review.
-
Synthetic Biological Circuits within an Orthogonal Central Dogma.Trends Biotechnol. 2021 Jan;39(1):59-71. doi: 10.1016/j.tibtech.2020.05.013. Epub 2020 Jun 22. Trends Biotechnol. 2021. PMID: 32586633 Free PMC article. Review.
-
Toehold switch based biosensors for sensing the highly trafficked rosewood Dalbergia maritima.Synth Syst Biotechnol. 2022 Mar 30;7(2):791-801. doi: 10.1016/j.synbio.2022.03.003. eCollection 2022 Jun. Synth Syst Biotechnol. 2022. PMID: 35415278 Free PMC article.
-
A versatile active learning workflow for optimization of genetic and metabolic networks.Nat Commun. 2022 Jul 5;13(1):3876. doi: 10.1038/s41467-022-31245-z. Nat Commun. 2022. PMID: 35790733 Free PMC article.
-
Synthetic biology and metabolic engineering for marine carotenoids: new opportunities and future prospects.Mar Drugs. 2014 Sep 17;12(9):4810-32. doi: 10.3390/md12094810. Mar Drugs. 2014. PMID: 25233369 Free PMC article. Review.
References
-
- Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. - PubMed
-
- Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. - PubMed
-
- Rinaudo K, Bleris L, Maddamsetti R, Subramanian S, Weiss R, Benenson Y. A universal RNAi-based logic evaluator that operates in mammalian cells. Nat. Biotechnol. 2007;25:795–801. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

