Transcriptional output in a prospective design conditionally on follow-up and exposure: the multistage model of cancer
- PMID: 22724047
- PMCID: PMC3376922
Transcriptional output in a prospective design conditionally on follow-up and exposure: the multistage model of cancer
Abstract
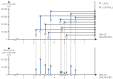
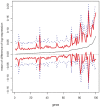
Transcriptomics as the analysis of mRNA and microRNA could be implemented in prospective studies both in peripheral blood and tissues. Its application in cancer epidemiology could provide a new understanding of the functional changes underlying the multistage model of carcinogenesis, as well as the relationship between these changes and exposure to carcinogens. Transcriptomics is not merely another -omics technology for risk assessment in traditional prospective studies. Instead, this novel approach has the potential to estimate the distribution of gene expression conditionally on different exposures, and to study the length of the different stages of carcinogenesis. If it proves to be a valid approach, transcriptomics could be an opportunity to make meaningful advances in our understanding of the carcinogenic process.
Keywords: Carcinogenesis; latent variable; multistage model; prospective study; systems epidemiology; transcriptomics.
Figures

Similar articles
-
A processual model for functional analyses of carcinogenesis in the prospective cohort design.Med Hypotheses. 2015 Oct;85(4):494-7. doi: 10.1016/j.mehy.2015.07.006. Epub 2015 Jul 11. Med Hypotheses. 2015. PMID: 26187155 Free PMC article.
-
An exposure driven functional model of carcinogenesis.Med Hypotheses. 2011 Aug;77(2):195-8. doi: 10.1016/j.mehy.2011.04.009. Epub 2011 May 6. Med Hypotheses. 2011. PMID: 21550177
-
Measuring the exposome: a powerful basis for evaluating environmental exposures and cancer risk.Environ Mol Mutagen. 2013 Aug;54(7):480-99. doi: 10.1002/em.21777. Epub 2013 May 16. Environ Mol Mutagen. 2013. PMID: 23681765 Review.
-
Towards a more functional concept of causality in cancer research.Int J Mol Epidemiol Genet. 2010 Mar 15;1(2):124-33. Int J Mol Epidemiol Genet. 2010. PMID: 21537385 Free PMC article.
-
Quantitative risk assessment and the limitations of the linearized multistage model.Hum Exp Toxicol. 1996 Feb;15(2):87-104. doi: 10.1177/096032719601500201. Hum Exp Toxicol. 1996. PMID: 8645508 Review.
Cited by
-
A processual model for functional analyses of carcinogenesis in the prospective cohort design.Med Hypotheses. 2015 Oct;85(4):494-7. doi: 10.1016/j.mehy.2015.07.006. Epub 2015 Jul 11. Med Hypotheses. 2015. PMID: 26187155 Free PMC article.
-
A new statistical method for curve group analysis of longitudinal gene expression data illustrated for breast cancer in the NOWAC postgenome cohort as a proof of principle.BMC Med Res Methodol. 2016 Mar 5;16:28. doi: 10.1186/s12874-016-0129-z. BMC Med Res Methodol. 2016. PMID: 26944545 Free PMC article.
-
Prediagnostic transcriptomic markers of Chronic lymphocytic leukemia reveal perturbations 10 years before diagnosis.Ann Oncol. 2014 May;25(5):1065-72. doi: 10.1093/annonc/mdu056. Epub 2014 Feb 20. Ann Oncol. 2014. PMID: 24558024 Free PMC article.
References
-
- Croce CM. Oncogenes and cancer. N Engl J Med. 2008;358:502–511. - PubMed
LinkOut - more resources
Full Text Sources
