Human α-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets
- PMID: 22722251
- PMCID: PMC4332406
- DOI: 10.1126/science.1218831
Human α-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets
Abstract
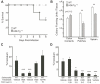
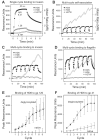
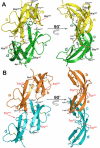
Defensins are antimicrobial peptides that contribute broadly to innate immunity, including protection of mucosal tissues. Human α-defensin (HD) 6 is highly expressed by secretory Paneth cells of the small intestine. However, in contrast to the other defensins, it lacks appreciable bactericidal activity. Nevertheless, we report here that HD6 affords protection against invasion by enteric bacterial pathogens in vitro and in vivo. After stochastic binding to bacterial surface proteins, HD6 undergoes ordered self-assembly to form fibrils and nanonets that surround and entangle bacteria. This self-assembly mechanism occurs in vivo, requires histidine-27, and is consistent with x-ray crystallography data. These findings support a key role for HD6 in protecting the small intestine against invasion by diverse enteric pathogens and may explain the conservation of HD6 throughout Hominidae evolution.
Figures

Comment in
-
Immunology. HD6 defensin nanonets.Science. 2012 Jul 27;337(6093):420-1. doi: 10.1126/science.1225906. Science. 2012. PMID: 22837514 No abstract available.
Similar articles
-
Paneth cell alpha-defensins: peptide mediators of innate immunity in the small intestine.Springer Semin Immunopathol. 2005 Sep;27(2):133-46. doi: 10.1007/s00281-005-0202-x. Epub 2005 Jun 2. Springer Semin Immunopathol. 2005. PMID: 15931529 Review.
-
Innate immune functions of α-defensins in the small intestine.Dig Dis. 2013;31(3-4):299-304. doi: 10.1159/000354681. Epub 2013 Nov 14. Dig Dis. 2013. PMID: 24246978 Review.
-
Human α-Defensin 6: A Small Peptide That Self-Assembles and Protects the Host by Entangling Microbes.Acc Chem Res. 2017 Apr 18;50(4):960-967. doi: 10.1021/acs.accounts.6b00653. Epub 2017 Mar 15. Acc Chem Res. 2017. PMID: 28296382 Free PMC article. Review.
-
Immunology. HD6 defensin nanonets.Science. 2012 Jul 27;337(6093):420-1. doi: 10.1126/science.1225906. Science. 2012. PMID: 22837514 No abstract available.
-
Paneth cells, defensins, and the commensal microbiota: a hypothesis on intimate interplay at the intestinal mucosa.Semin Immunol. 2007 Apr;19(2):70-83. doi: 10.1016/j.smim.2007.04.002. Epub 2007 May 7. Semin Immunol. 2007. PMID: 17485224 Review.
Cited by
-
Oh what tangled webs we weave….Gut Microbes. 2013 May-Jun;4(3):179-80. doi: 10.4161/gmic.24388. Epub 2013 Mar 21. Gut Microbes. 2013. PMID: 23519061 Free PMC article.
-
Inflammatory bowel disease: an impaired barrier disease.Langenbecks Arch Surg. 2013 Jan;398(1):1-12. doi: 10.1007/s00423-012-1030-9. Epub 2012 Nov 18. Langenbecks Arch Surg. 2013. PMID: 23160753 Review.
-
Chronic Heavy Alcohol Use is Associated with Upregulated Paneth Cell Antimicrobials in Gastric Mucosa.Clin Transl Gastroenterol. 2015 Jul 16;6(7):e103. doi: 10.1038/ctg.2015.26. Clin Transl Gastroenterol. 2015. PMID: 26181292 Free PMC article.
-
Human intelectin-2 (ITLN2) is selectively expressed by secretory Paneth cells.FASEB J. 2022 Mar;36(3):e22200. doi: 10.1096/fj.202101870R. FASEB J. 2022. PMID: 35182405 Free PMC article.
-
Mechanisms and regulation of defensins in host defense.Signal Transduct Target Ther. 2023 Aug 14;8(1):300. doi: 10.1038/s41392-023-01553-x. Signal Transduct Target Ther. 2023. PMID: 37574471 Free PMC article. Review.
References
-
- Lehrer RI, Bevins CL, Ganz T. In: Mucosal Immunology. 3rd Mestecky J, et al., editors. Vol. 1. Academic Press; New York: 2004. pp. 95–110.
-
- Ouellette AJ. Paneth cells and innate mucosal immunity. Curr Opin Gastroenterol. 2010;26:547. - PubMed
-
- Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011;9:356. - PubMed
-
- Wehkamp J, Stange EF. Paneth's disease. J. Crohns Colitis. 2010;4:523. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI082320/AI/NIAID NIH HHS/United States
- T32 AI060555/AI/NIAID NIH HHS/United States
- R01 AI044170/AI/NIAID NIH HHS/United States
- AI050843/AI/NIAID NIH HHS/United States
- R21 AI057757/AI/NIAID NIH HHS/United States
- R21 AI073120/AI/NIAID NIH HHS/United States
- AI070726/AI/NIAID NIH HHS/United States
- AI040124/AI/NIAID NIH HHS/United States
- HD059127/HD/NICHD NIH HHS/United States
- R21 AI088122/AI/NIAID NIH HHS/United States
- R01 AI076246/AI/NIAID NIH HHS/United States
- AI073120/AI/NIAID NIH HHS/United States
- R29 AI040124/AI/NIAID NIH HHS/United States
- R01 AI070726/AI/NIAID NIH HHS/United States
- R01 AI032738/AI/NIAID NIH HHS/United States
- R01 AI072732/AI/NIAID NIH HHS/United States
- R01 AI040124/AI/NIAID NIH HHS/United States
- T32AI060555/AI/NIAID NIH HHS/United States
- AI088122/AI/NIAID NIH HHS/United States
- AI076246/AI/NIAID NIH HHS/United States
- R21 AI082320/AI/NIAID NIH HHS/United States
- AI072732/AI/NIAID NIH HHS/United States
- AI032738/AI/NIAID NIH HHS/United States
- R37 AI032738/AI/NIAID NIH HHS/United States
- R01 AI057757/AI/NIAID NIH HHS/United States
- R01 AI050843/AI/NIAID NIH HHS/United States
- AI044170/AI/NIAID NIH HHS/United States
- AI057757/AI/NIAID NIH HHS/United States
- R01 GM099526/GM/NIGMS NIH HHS/United States
- R01 HD059127/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

