The early UL31 gene of equine herpesvirus 1 encodes a single-stranded DNA-binding protein that has a nuclear localization signal sequence at the C-terminus
- PMID: 22721961
- PMCID: PMC3423489
- DOI: 10.1016/j.virol.2012.05.031
The early UL31 gene of equine herpesvirus 1 encodes a single-stranded DNA-binding protein that has a nuclear localization signal sequence at the C-terminus
Abstract
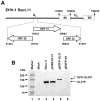
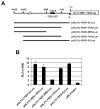
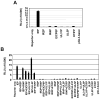
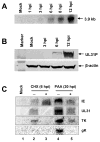
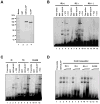
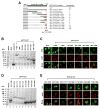
The amino acid sequence of the UL31 protein (UL31P) of equine herpesvirus 1 (EHV-1) has homology to that of the ICP8 of herpes simplex virus type 1 (HSV-1). Here we show that the UL31 gene is synergistically trans-activated by the IEP and the UL5P (EICP27). Detection of the UL31 RNA transcript and the UL31P in EHV-1-infected cells at 6h post-infection (hpi) as well as metabolic inhibition assays indicated that UL31 is an early gene. The UL31P preferentially bound to single-stranded DNA over double-stranded DNA in gel shift assays. Subcellular localization of the green fluorescent protein (GFP)-UL31 fusion proteins revealed that the C-terminal 32 amino acid residues of the UL31P are responsible for the nuclear localization. These findings may contribute to defining the role of the UL31P single-stranded DNA-binding protein in EHV-1 DNA replication.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Cloning and expression of an equine herpesvirus 1 origin-binding protein.J Virol. 1994 Jun;68(6):3674-81. doi: 10.1128/JVI.68.6.3674-3681.1994. J Virol. 1994. PMID: 8189505 Free PMC article.
-
Characterization of the trans-activation properties of equine herpesvirus 1 EICP0 protein.J Virol. 2000 Feb;74(3):1200-8. doi: 10.1128/jvi.74.3.1200-1208.2000. J Virol. 2000. PMID: 10627530 Free PMC article.
-
Differences in determinants required for complex formation and transactivation in related VP16 proteins.J Virol. 2000 Nov;74(21):10112-21. doi: 10.1128/jvi.74.21.10112-10121.2000. J Virol. 2000. PMID: 11024140 Free PMC article.
-
Identification of an essential domain in the herpes simplex virus 1 UL34 protein that is necessary and sufficient to interact with UL31 protein.J Virol. 2005 Mar;79(6):3797-806. doi: 10.1128/JVI.79.6.3797-3806.2005. J Virol. 2005. PMID: 15731273 Free PMC article.
-
Characterization of the nuclear import and export signals of pseudorabies virus UL31.Arch Virol. 2015 Oct;160(10):2591-4. doi: 10.1007/s00705-015-2527-7. Epub 2015 Jul 21. Arch Virol. 2015. PMID: 26195191
Cited by
-
Comparative Genomic Sequencing and Pathogenic Properties of Equine Herpesvirus 1 KyA and RacL11.Front Vet Sci. 2017 Dec 11;4:211. doi: 10.3389/fvets.2017.00211. eCollection 2017. Front Vet Sci. 2017. PMID: 29312962 Free PMC article.
-
Regulation of alphaherpesvirus protein via post-translational phosphorylation.Vet Res. 2022 Nov 17;53(1):93. doi: 10.1186/s13567-022-01115-z. Vet Res. 2022. PMID: 36397147 Free PMC article. Review.
-
Interactions of the Kaposi's Sarcoma-associated herpesvirus nuclear egress complex: ORF69 is a potent factor for remodeling cellular membranes.J Virol. 2013 Apr;87(7):3915-29. doi: 10.1128/JVI.03418-12. Epub 2013 Jan 30. J Virol. 2013. PMID: 23365436 Free PMC article.
References
-
- Allen GP, Bryans JT. Molecular epizootiology, pathogenesis, and prophylaxis of equine herpesvirus-1 infections. Prog Vet Microbiol Immunol. 1986;2:78–144. - PubMed
-
- Allen GP, O’Callaghan DJ, Randall CC. Purification and characterization of equine herpesvirus-induced DNA polymerase. Virology. 1977;76:395–408. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

