eIF4E-binding protein regulation of mRNAs with differential 5'-UTR secondary structure: a polyelectrostatic model for a component of protein-mRNA interactions
- PMID: 22718971
- PMCID: PMC3439904
- DOI: 10.1093/nar/gks511
eIF4E-binding protein regulation of mRNAs with differential 5'-UTR secondary structure: a polyelectrostatic model for a component of protein-mRNA interactions
Abstract
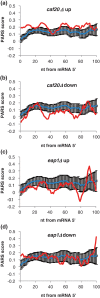
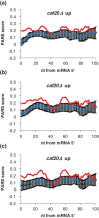
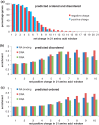
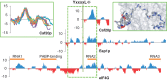
Control of translation in eukaryotes is complex, depending on the binding of various factors to mRNAs. Available data for subsets of mRNAs that are translationally up- and down-regulated in yeast eIF4E-binding protein (4E-BP) deletion mutants are coupled with reported mRNA secondary structure measurements to investigate whether 5'-UTR secondary structure varies between the subsets. Genes with up-regulated translational efficiencies in the caf20Δ mutant have relatively high averaged 5'-UTR secondary structure. There is no apparent wide-scale correlation of RNA-binding protein preferences with the increased 5'-UTR secondary structure, leading us to speculate that the secondary structure itself may play a role in differential partitioning of mRNAs between eIF4E/4E-BP repression and eIF4E/eIF4G translation initiation. Both Caf20p and Eap1p contain stretches of positive charge in regions of predicted disorder. Such regions are also present in eIF4G and have been reported to associate with mRNA binding. The pattern of these segments, around the canonical eIF4E-binding motif, varies between each 4E-BP and eIF4G. Analysis of gene ontology shows that yeast proteins containing predicted disordered segments, with positive charge runs, are enriched for nucleic acid binding. We propose that the 4E-BPs act, in part, as differential, flexible, polyelectrostatic scaffolds for mRNAs.
Figures

Similar articles
-
Structural motifs in eIF4G and 4E-BPs modulate their binding to eIF4E to regulate translation initiation in yeast.Nucleic Acids Res. 2018 Jul 27;46(13):6893-6908. doi: 10.1093/nar/gky542. Nucleic Acids Res. 2018. PMID: 30053226 Free PMC article.
-
The 4E-BP Caf20p Mediates Both eIF4E-Dependent and Independent Repression of Translation.PLoS Genet. 2015 May 14;11(5):e1005233. doi: 10.1371/journal.pgen.1005233. eCollection 2015 May. PLoS Genet. 2015. PMID: 25973932 Free PMC article.
-
eIF4E and Interactors from Unicellular Eukaryotes.Int J Mol Sci. 2020 Mar 21;21(6):2170. doi: 10.3390/ijms21062170. Int J Mol Sci. 2020. PMID: 32245232 Free PMC article. Review.
-
Identifying eIF4E-binding protein translationally-controlled transcripts reveals links to mRNAs bound by specific PUF proteins.Nucleic Acids Res. 2010 Dec;38(22):8039-50. doi: 10.1093/nar/gkq686. Epub 2010 Aug 12. Nucleic Acids Res. 2010. PMID: 20705650 Free PMC article.
-
Translational control of mRNAs by 3'-Untranslated region binding proteins.BMB Rep. 2017 Apr;50(4):194-200. doi: 10.5483/bmbrep.2017.50.4.040. BMB Rep. 2017. PMID: 28287067 Free PMC article. Review.
Cited by
-
Integrin β1 mediates 5-fluorouracil chemoresistance under translational control of eIF4E in colorectal cancer.Int J Clin Exp Pathol. 2018 Oct 1;11(10):4771-4783. eCollection 2018. Int J Clin Exp Pathol. 2018. PMID: 31949552 Free PMC article.
-
Soluble expression of proteins correlates with a lack of positively-charged surface.Sci Rep. 2013 Nov 26;3:3333. doi: 10.1038/srep03333. Sci Rep. 2013. PMID: 24276756 Free PMC article.
-
Structural motifs in eIF4G and 4E-BPs modulate their binding to eIF4E to regulate translation initiation in yeast.Nucleic Acids Res. 2018 Jul 27;46(13):6893-6908. doi: 10.1093/nar/gky542. Nucleic Acids Res. 2018. PMID: 30053226 Free PMC article.
-
The 4E-BP Caf20p Mediates Both eIF4E-Dependent and Independent Repression of Translation.PLoS Genet. 2015 May 14;11(5):e1005233. doi: 10.1371/journal.pgen.1005233. eCollection 2015 May. PLoS Genet. 2015. PMID: 25973932 Free PMC article.
-
eIF4E and Interactors from Unicellular Eukaryotes.Int J Mol Sci. 2020 Mar 21;21(6):2170. doi: 10.3390/ijms21062170. Int J Mol Sci. 2020. PMID: 32245232 Free PMC article. Review.
References
-
- Raught B, Gingras AC. eIF4E activity is regulated at multiple levels. Int. J. Biochem. Cell Biol. 1999;31:43–57. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

