Adenoviral protein V promotes a process of viral assembly through nucleophosmin 1
- PMID: 22717133
- PMCID: PMC3423539
- DOI: 10.1016/j.virol.2012.05.028
Adenoviral protein V promotes a process of viral assembly through nucleophosmin 1
Abstract
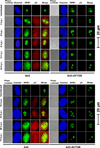
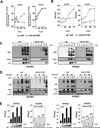
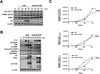
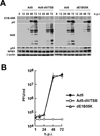
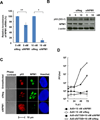
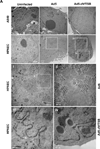
Adenoviral infection induces nucleoplasmic redistribution of a nucleolar nucleophosmin 1/NPM1/B23.1. NPM1 is preferentially localized in the nucleoli of normal cells, whereas it is also present at the nuclear matrix in cancer cells. However, the biological roles of NPM1 during infection are unknown. Here, by analyzing a pV-deletion mutant, Ad5-dV/TSB, we demonstrate that pV promotes the NPM1 translocation from the nucleoli to the nucleoplasm in normal cells, and the NPM1 translocation is correlated with adenoviral replication. Lack of pV causes a dramatic reduction of adenoviral replication in normal cells, but not cancer cells, and Ad5-dV/TSB was defective in viral assembly in normal cells. NPM1 knockdown inhibits adenoviral replication, suggesting an involvement of NPM1 in adenoviral biology. Further, we show that NPM1 interacts with empty adenovirus particles which are an intermediate during virion maturation by immunoelectron microscopy. Collectively, these data implicate that pV participates in a process of viral assembly through NPM1.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Nuclear reorganization by NPM1-mediated phase separation triggered by adenovirus core protein VII.Microbiol Spectr. 2024 Oct 3;12(10):e0041624. doi: 10.1128/spectrum.00416-24. Epub 2024 Aug 20. Microbiol Spectr. 2024. PMID: 39162498 Free PMC article.
-
Nucleolar protein B23 interacts with Japanese encephalitis virus core protein and participates in viral replication.Microbiol Immunol. 2006;50(3):225-34. doi: 10.1111/j.1348-0421.2006.tb03789.x. Microbiol Immunol. 2006. PMID: 16547420
-
Identification of nucleophosmin/B23, an acidic nucleolar protein, as a stimulatory factor for in vitro replication of adenovirus DNA complexed with viral basic core proteins.J Mol Biol. 2001 Aug 3;311(1):41-55. doi: 10.1006/jmbi.2001.4812. J Mol Biol. 2001. PMID: 11469856
-
Implication of B23/NPM1 in Viral Infections, Potential Uses of B23/NPM1 Inhibitors as Antiviral Therapy.Infect Disord Drug Targets. 2019;19(1):2-16. doi: 10.2174/1871526518666180327124412. Infect Disord Drug Targets. 2019. PMID: 29589547 Review.
-
Nucleophosmin in Its Interaction with Ligands.Int J Mol Sci. 2020 Jul 10;21(14):4885. doi: 10.3390/ijms21144885. Int J Mol Sci. 2020. PMID: 32664415 Free PMC article. Review.
Cited by
-
Adenoviral vector-based strategies against infectious disease and cancer.Hum Vaccin Immunother. 2016 Aug 2;12(8):2064-2074. doi: 10.1080/21645515.2016.1165908. Epub 2016 Apr 22. Hum Vaccin Immunother. 2016. PMID: 27105067 Free PMC article. Review.
-
RANBP2 and USP9x regulate nuclear import of adenovirus minor coat protein IIIa.PLoS Pathog. 2022 Jun 16;18(6):e1010588. doi: 10.1371/journal.ppat.1010588. eCollection 2022 Jun. PLoS Pathog. 2022. PMID: 35709296 Free PMC article.
-
Lytic Reactivation of the Kaposi's Sarcoma-Associated Herpesvirus (KSHV) Is Accompanied by Major Nucleolar Alterations.Viruses. 2022 Aug 4;14(8):1720. doi: 10.3390/v14081720. Viruses. 2022. PMID: 36016343 Free PMC article.
-
Species D human adenovirus type 9 exhibits better virus-spread ability for antitumor efficacy among alternative serotypes.PLoS One. 2014 Feb 4;9(2):e87342. doi: 10.1371/journal.pone.0087342. eCollection 2014. PLoS One. 2014. PMID: 24503714 Free PMC article.
-
The function of DNA binding protein nucleophosmin in AAV replication.Virology. 2017 Oct;510:46-54. doi: 10.1016/j.virol.2017.07.007. Epub 2017 Jul 10. Virology. 2017. PMID: 28704696 Free PMC article.
References
-
- Anderson CW, Young ME, Flint SJ. Characterization of the adenovirus 2 virion protein, mu. Virology. 1989;172(2):506–512. - PubMed
-
- Barker DD, Berk AJ. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology. 1987;156(1):107–121. - PubMed
-
- Bischoff JR, Kirn DH, Williams A, Heise C, Horn S, Muna M, Ng L, Nye JA, Sampson-Johannes A, Fattaey A, McCormick F. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science. 1996;274(5286):373–376. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

