Structure of p22 headful packaging nuclease
- PMID: 22715098
- PMCID: PMC3431676
- DOI: 10.1074/jbc.M112.349894
Structure of p22 headful packaging nuclease
Abstract
Packaging of viral genomes into preformed procapsids requires the controlled and synchronized activity of an ATPase and a genome-processing nuclease, both located in the large terminase (L-terminase) subunit. In this paper, we have characterized the structure and regulation of bacteriophage P22 L-terminase (gp2). Limited proteolysis reveals a bipartite organization consisting of an N-terminal ATPase core flexibly connected to a C-terminal nuclease domain. The 2.02 Å crystal structure of P22 headful nuclease obtained by in-drop proteolysis of full-length L-terminase (FL-L-terminase) reveals a central seven-stranded β-sheet core that harbors two magnesium ions. Modeling studies with DNA suggest that the two ions are poised for two-metal ion-dependent catalysis, but the nuclease DNA binding surface is sterically hindered by a loop-helix (L(1)-α(2)) motif, which is incompatible with catalysis. Accordingly, the isolated nuclease is completely inactive in vitro, whereas it exhibits endonucleolytic activity in the context of FL-L-terminase. Deleting the autoinhibitory L(1)-α(2) motif (or just the loop L(1)) restores nuclease activity to a level comparable with FL-L-terminase. Together, these results suggest that the activity of P22 headful nuclease is regulated by intramolecular cross-talk with the N-terminal ATPase domain. This cross-talk allows for precise and controlled cleavage of DNA that is essential for genome packaging.
Figures

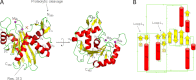
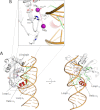

Similar articles
-
The headful packaging nuclease of bacteriophage T4.Mol Microbiol. 2008 Sep;69(5):1180-90. doi: 10.1111/j.1365-2958.2008.06344.x. Epub 2008 Jul 4. Mol Microbiol. 2008. PMID: 18627466
-
Small terminase couples viral DNA binding to genome-packaging ATPase activity.Structure. 2012 Aug 8;20(8):1403-13. doi: 10.1016/j.str.2012.05.014. Epub 2012 Jul 5. Structure. 2012. PMID: 22771211 Free PMC article.
-
Architecture of the Complex Formed by Large and Small Terminase Subunits from Bacteriophage P22.J Mol Biol. 2015 Oct 9;427(20):3285-3299. doi: 10.1016/j.jmb.2015.08.013. Epub 2015 Aug 21. J Mol Biol. 2015. PMID: 26301600 Free PMC article.
-
Terminase Large Subunit Provides a New Drug Target for Herpesvirus Treatment.Viruses. 2019 Mar 5;11(3):219. doi: 10.3390/v11030219. Viruses. 2019. PMID: 30841485 Free PMC article. Review.
-
Headful DNA packaging: bacteriophage SPP1 as a model system.Virus Res. 2013 May;173(2):247-59. doi: 10.1016/j.virusres.2013.01.021. Epub 2013 Feb 16. Virus Res. 2013. PMID: 23419885 Review.
Cited by
-
Structures of the phage Sf6 large terminase provide new insights into DNA translocation and cleavage.Proc Natl Acad Sci U S A. 2013 May 14;110(20):8075-80. doi: 10.1073/pnas.1301133110. Epub 2013 Apr 29. Proc Natl Acad Sci U S A. 2013. PMID: 23630261 Free PMC article.
-
The enzymology of a viral genome packaging motor is influenced by the assembly state of the motor subunits.Biochemistry. 2012 Nov 20;51(46):9342-53. doi: 10.1021/bi300890y. Epub 2012 Nov 7. Biochemistry. 2012. PMID: 23134123 Free PMC article.
-
Recognition of an α-helical hairpin in P22 large terminase by a synthetic antibody fragment.Acta Crystallogr D Struct Biol. 2020 Sep 1;76(Pt 9):876-888. doi: 10.1107/S2059798320009912. Epub 2020 Aug 17. Acta Crystallogr D Struct Biol. 2020. PMID: 32876063 Free PMC article.
-
Bacteriophage T4 Head: Structure, Assembly, and Genome Packaging.Viruses. 2023 Feb 14;15(2):527. doi: 10.3390/v15020527. Viruses. 2023. PMID: 36851741 Free PMC article. Review.
-
Physical and Functional Characterization of a Viral Genome Maturation Complex.Biophys J. 2017 Apr 25;112(8):1551-1560. doi: 10.1016/j.bpj.2017.02.041. Biophys J. 2017. PMID: 28445747 Free PMC article.
References
-
- Casjens S. R. (2011) The DNA packaging nanomotor of tailed bacteriophages. Nat. Rev. Microbiol. 9, 647–657 - PubMed
-
- Rao V. B., Feiss M. (2008) The bacteriophage DNA packaging motor. Annu. Rev. Genet. 42, 647–681 - PubMed
-
- Sun S., Kondabagil K., Gentz P. M., Rossmann M. G., Rao V. B. (2007) The structure of the ATPase that powers DNA packaging into bacteriophage T4 procapsids. Mol. Cell 25, 943–949 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

