Nuclear envelope morphology constrains diffusion and promotes asymmetric protein segregation in closed mitosis
- PMID: 22711697
- PMCID: PMC3384416
- DOI: 10.1083/jcb.201112117
Nuclear envelope morphology constrains diffusion and promotes asymmetric protein segregation in closed mitosis
Erratum in
- J Cell Biol. 2012 Jul 9;198(1):143
Abstract
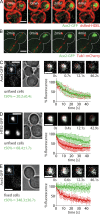
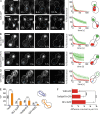
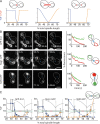
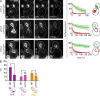
During vegetative growth, Saccharomyces cerevisiae cells divide asymmetrically: the mother cell buds to produce a smaller daughter cell. This daughter asymmetrically inherits the transcription factor Ace2, which activates daughter-specific transcriptional programs. In this paper, we investigate when and how this asymmetry is established and maintained. We show that Ace2 asymmetry is initiated in the elongated, but undivided, anaphase nucleus. At this stage, the nucleoplasm was highly compartmentalized; little exchange was observed for nucleoplasmic proteins between mother and bud. Using photobleaching and in silico modeling, we show that diffusion barriers compartmentalize the nuclear membranes. In contrast, the behavior of proteins in the nucleoplasm is well explained by the dumbbell shape of the anaphase nucleus. This compartmentalization of the nucleoplasm promoted Ace2 asymmetry in anaphase nuclei. Thus, our data indicate that yeast cells use the process of closed mitosis and the morphological constraints associated with it to asymmetrically segregate nucleoplasmic components.
Figures

Similar articles
-
A mechanism for asymmetric segregation of age during yeast budding.Nature. 2008 Aug 7;454(7205):728-34. doi: 10.1038/nature07212. Epub 2008 Jul 27. Nature. 2008. PMID: 18660802
-
Asymmetric Transcription Factor Partitioning During Yeast Cell Division Requires the FACT Chromatin Remodeler and Cell Cycle Progression.Genetics. 2020 Nov;216(3):701-716. doi: 10.1534/genetics.120.303439. Epub 2020 Sep 2. Genetics. 2020. PMID: 32878900 Free PMC article.
-
The NDR/LATS family kinase Cbk1 directly controls transcriptional asymmetry.PLoS Biol. 2008 Aug 19;6(8):e203. doi: 10.1371/journal.pbio.0060203. PLoS Biol. 2008. PMID: 18715118 Free PMC article.
-
Mitotic exit and separation of mother and daughter cells.Genetics. 2012 Dec;192(4):1165-202. doi: 10.1534/genetics.112.145516. Genetics. 2012. PMID: 23212898 Free PMC article. Review.
-
Daughter-specific repression of Saccharomyces cerevisiae HO: Ash1 is the commander.EMBO Rep. 2004 Oct;5(10):953-7. doi: 10.1038/sj.embor.7400251. EMBO Rep. 2004. PMID: 15459746 Free PMC article. Review.
Cited by
-
Asymmetric nuclear division in neural stem cells generates sibling nuclei that differ in size, envelope composition, and chromatin organization.Curr Biol. 2021 Sep 27;31(18):3973-3983.e4. doi: 10.1016/j.cub.2021.06.063. Epub 2021 Jul 22. Curr Biol. 2021. PMID: 34297912 Free PMC article.
-
DNA circles promote yeast ageing in part through stimulating the reorganization of nuclear pore complexes.Elife. 2022 Apr 4;11:e71196. doi: 10.7554/eLife.71196. Elife. 2022. PMID: 35373738 Free PMC article.
-
A sphingolipid-dependent diffusion barrier confines ER stress to the yeast mother cell.Elife. 2014 May 6;3:e01883. doi: 10.7554/eLife.01883. Elife. 2014. PMID: 24843009 Free PMC article.
-
Budding yeast Wee1 distinguishes spindle pole bodies to guide their pattern of age-dependent segregation.Nat Cell Biol. 2017 Aug;19(8):941-951. doi: 10.1038/ncb3576. Epub 2017 Jul 17. Nat Cell Biol. 2017. PMID: 28714971
-
Mixing and matching nuclear envelope remodeling and spindle assembly strategies in the evolution of mitosis.Curr Opin Cell Biol. 2016 Aug;41:43-50. doi: 10.1016/j.ceb.2016.03.016. Epub 2016 Apr 7. Curr Opin Cell Biol. 2016. PMID: 27062548 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

