Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction
- PMID: 22708584
- PMCID: PMC3412702
- DOI: 10.1186/1471-2105-13-134
Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction
Abstract
Background: Choosing appropriate primers is probably the single most important factor affecting the polymerase chain reaction (PCR). Specific amplification of the intended target requires that primers do not have matches to other targets in certain orientations and within certain distances that allow undesired amplification. The process of designing specific primers typically involves two stages. First, the primers flanking regions of interest are generated either manually or using software tools; then they are searched against an appropriate nucleotide sequence database using tools such as BLAST to examine the potential targets. However, the latter is not an easy process as one needs to examine many details between primers and targets, such as the number and the positions of matched bases, the primer orientations and distance between forward and reverse primers. The complexity of such analysis usually makes this a time-consuming and very difficult task for users, especially when the primers have a large number of hits. Furthermore, although the BLAST program has been widely used for primer target detection, it is in fact not an ideal tool for this purpose as BLAST is a local alignment algorithm and does not necessarily return complete match information over the entire primer range.
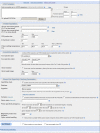
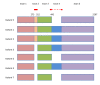
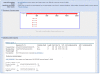
Results: We present a new software tool called Primer-BLAST to alleviate the difficulty in designing target-specific primers. This tool combines BLAST with a global alignment algorithm to ensure a full primer-target alignment and is sensitive enough to detect targets that have a significant number of mismatches to primers. Primer-BLAST allows users to design new target-specific primers in one step as well as to check the specificity of pre-existing primers. Primer-BLAST also supports placing primers based on exon/intron locations and excluding single nucleotide polymorphism (SNP) sites in primers.
Conclusions: We describe a robust and fully implemented general purpose primer design tool that designs target-specific PCR primers. Primer-BLAST offers flexible options to adjust the specificity threshold and other primer properties. This tool is publicly available at http://www.ncbi.nlm.nih.gov/tools/primer-blast.
Figures


Similar articles
-
ConservedPrimers 2.0: a high-throughput pipeline for comparative genome referenced intron-flanking PCR primer design and its application in wheat SNP discovery.BMC Bioinformatics. 2009 Oct 13;10:331. doi: 10.1186/1471-2105-10-331. BMC Bioinformatics. 2009. PMID: 19825183 Free PMC article.
-
BatchPrimer3: a high throughput web application for PCR and sequencing primer design.BMC Bioinformatics. 2008 May 29;9:253. doi: 10.1186/1471-2105-9-253. BMC Bioinformatics. 2008. PMID: 18510760 Free PMC article.
-
RExPrimer: an integrated primer designing tool increases PCR effectiveness by avoiding 3' SNP-in-primer and mis-priming from structural variation.BMC Genomics. 2009 Dec 3;10 Suppl 3(Suppl 3):S4. doi: 10.1186/1471-2164-10-S3-S4. BMC Genomics. 2009. PMID: 19958502 Free PMC article.
-
Graphical design of primers with PerlPrimer.Methods Mol Biol. 2007;402:403-14. doi: 10.1007/978-1-59745-528-2_21. Methods Mol Biol. 2007. PMID: 17951808 Review.
-
PCR primer design using statistical modeling.Methods Mol Biol. 2007;402:93-104. doi: 10.1007/978-1-59745-528-2_5. Methods Mol Biol. 2007. PMID: 17951792 Review.
Cited by
-
Integrating molecular, biochemical, and immunohistochemical features as predictors of hepatocellular carcinoma drug response using machine-learning algorithms.Front Mol Biosci. 2024 Oct 16;11:1430794. doi: 10.3389/fmolb.2024.1430794. eCollection 2024. Front Mol Biosci. 2024. PMID: 39479501 Free PMC article.
-
Ancestral lineages of dietary exposure to an endocrine disrupting chemical drive distinct forms of transgenerational subfertility in an insect model.Sci Rep. 2024 Aug 5;14(1):18153. doi: 10.1038/s41598-024-67921-x. Sci Rep. 2024. PMID: 39103404 Free PMC article.
-
Expression of Toll-like receptor 2, Dectin-1, and Osteopontin in murine model of pulpitis.Clin Oral Investig. 2023 Mar;27(3):1177-1192. doi: 10.1007/s00784-022-04732-2. Epub 2022 Oct 7. Clin Oral Investig. 2023. PMID: 36205788
-
Development of a mRNA profiling multiplex for the inference of organ tissues.Int J Legal Med. 2013 Sep;127(5):891-900. doi: 10.1007/s00414-013-0895-7. Epub 2013 Jul 10. Int J Legal Med. 2013. PMID: 23839651
-
A triple-masked, two-center, randomized parallel clinical trial to assess the superiority of eight weeks of grape seed flour supplementation against placebo for weight loss attenuation during perioperative period in patients with cachexia associated with colorectal cancer: a study protocol.Front Endocrinol (Lausanne). 2024 Jan 19;14:1146479. doi: 10.3389/fendo.2023.1146479. eCollection 2023. Front Endocrinol (Lausanne). 2024. PMID: 38313843 Free PMC article.
References
-
- VanGuilder HD, Vrana KE, Freeman WM. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques. 2008;44(5):619–626. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

