Human gut microbiome viewed across age and geography
- PMID: 22699611
- PMCID: PMC3376388
- DOI: 10.1038/nature11053
Human gut microbiome viewed across age and geography
Abstract
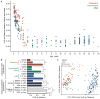
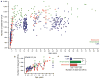
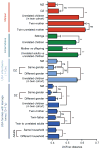
Gut microbial communities represent one source of human genetic and metabolic diversity. To examine how gut microbiomes differ among human populations, here we characterize bacterial species in fecal samples from 531 individuals, plus the gene content of 110 of them. The cohort encompassed healthy children and adults from the Amazonas of Venezuela, rural Malawi and US metropolitan areas and included mono- and dizygotic twins. Shared features of the functional maturation of the gut microbiome were identified during the first three years of life in all three populations, including age-associated changes in the genes involved in vitamin biosynthesis and metabolism. Pronounced differences in bacterial assemblages and functional gene repertoires were noted between US residents and those in the other two countries. These distinctive features are evident in early infancy as well as adulthood. Our findings underscore the need to consider the microbiome when evaluating human development, nutritional needs, physiological variations and the impact of westernization.
Figures

Similar articles
-
A core gut microbiome in obese and lean twins.Nature. 2009 Jan 22;457(7228):480-4. doi: 10.1038/nature07540. Epub 2008 Nov 30. Nature. 2009. PMID: 19043404 Free PMC article.
-
Distinct distal gut microbiome diversity and composition in healthy children from Bangladesh and the United States.PLoS One. 2013;8(1):e53838. doi: 10.1371/journal.pone.0053838. Epub 2013 Jan 22. PLoS One. 2013. PMID: 23349750 Free PMC article.
-
Distinct cutaneous bacterial assemblages in a sampling of South American Amerindians and US residents.ISME J. 2013 Jan;7(1):85-95. doi: 10.1038/ismej.2012.81. Epub 2012 Aug 16. ISME J. 2013. PMID: 22895161 Free PMC article.
-
Microbiome Signatures Associated With Steatohepatitis and Moderate to Severe Fibrosis in Children With Nonalcoholic Fatty Liver Disease.Gastroenterology. 2019 Oct;157(4):1109-1122. doi: 10.1053/j.gastro.2019.06.028. Epub 2019 Jun 27. Gastroenterology. 2019. PMID: 31255652 Free PMC article.
-
Comparative fecal metagenomics unveils unique functional capacity of the swine gut.BMC Microbiol. 2011 May 15;11:103. doi: 10.1186/1471-2180-11-103. BMC Microbiol. 2011. PMID: 21575148 Free PMC article.
Cited by
-
Diagnosing and engineering gut microbiomes.EMBO Mol Med. 2024 Oct 28. doi: 10.1038/s44321-024-00149-4. Online ahead of print. EMBO Mol Med. 2024. PMID: 39468301 Review.
-
Gut microbiota dysbiosis-mediated ceramides elevation contributes to corticosterone-induced depression by impairing mitochondrial function.NPJ Biofilms Microbiomes. 2024 Oct 28;10(1):111. doi: 10.1038/s41522-024-00582-w. NPJ Biofilms Microbiomes. 2024. PMID: 39468065 Free PMC article.
-
Association Between Gut Microbiome Composition and Physical Characteristics in Patients with Severe Motor and Intellectual Disabilities: Perspectives from Microbial Diversity.Nutrients. 2024 Oct 19;16(20):3546. doi: 10.3390/nu16203546. Nutrients. 2024. PMID: 39458540 Free PMC article.
-
Impact of Bifidobacterium longum Subspecies infantis on Pediatric Gut Health and Nutrition: Current Evidence and Future Directions.Nutrients. 2024 Oct 16;16(20):3510. doi: 10.3390/nu16203510. Nutrients. 2024. PMID: 39458503 Free PMC article. Review.
-
Oral Microbial Translocation Genes in Gastrointestinal Cancers: Insights from Metagenomic Analysis.Microorganisms. 2024 Oct 18;12(10):2086. doi: 10.3390/microorganisms12102086. Microorganisms. 2024. PMID: 39458395 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

