Variation of presence/absence genes among Arabidopsis populations
- PMID: 22697058
- PMCID: PMC3433342
- DOI: 10.1186/1471-2148-12-86
Variation of presence/absence genes among Arabidopsis populations
Abstract
Background: Gene presence/absence (P/A) polymorphisms are commonly observed in plants and are important in individual adaptation and species differentiation. Detecting their abundance, distribution and variation among individuals would help to understand the role played by these polymorphisms in a given species. The recently sequenced 80 Arabidopsis genomes provide an opportunity to address these questions.
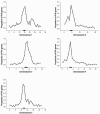
Results: By systematically investigating these accessions, we identified 2,407 P/A genes (or 8.9%) absent in one or more genomes, averaging 444 absent genes per accession. 50.6% of P/A genes belonged to multi-copy gene families, or 31.0% to clustered genes. However, the highest proportion of P/A genes, outnumbered in singleton genes, was observed in the regions near centromeres. In addition, a significant correlation was observed between the P/A gene frequency among the 80 accessions and the diversity level at P/A loci. Furthermore, the proportion of P/A genes was different among functional gene categories. Finally, a P/A gene tree showed a diversified population structure in the worldwide Arabidopsis accessions.
Conclusions: An estimate of P/A genes and their frequency distribution in the worldwide Arabidopsis accessions was obtained. Our results suggest that there are diverse mechanisms to generate or maintain P/A genes, by which individuals and functionally different genes can selectively maintain P/A polymorphisms for a specific adaptation.
Figures
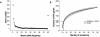
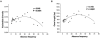


Similar articles
-
Grouped nucleotide polymorphism: a major contributor to genetic variation in Arabidopsis.Gene. 2008 Dec 15;426(1-2):1-6. doi: 10.1016/j.gene.2008.09.003. Epub 2008 Sep 12. Gene. 2008. PMID: 18835338
-
Unique evolutionary mechanism in R-genes under the presence/absence polymorphism in Arabidopsis thaliana.Genetics. 2006 Feb;172(2):1243-50. doi: 10.1534/genetics.105.047290. Epub 2006 Feb 1. Genetics. 2006. PMID: 16452149 Free PMC article.
-
Transposon-associated polymorphisms of stress-responsive gene promoters in selected accessions of Arabidopsis thaliana.Acta Biochim Pol. 2018;65(3):391-396. doi: 10.18388/abp.2017_1590. Epub 2018 Aug 27. Acta Biochim Pol. 2018. PMID: 30148504
-
From phenotypic to molecular polymorphisms involved in naturally occurring variation of plant development.Int J Dev Biol. 2005;49(5-6):717-32. doi: 10.1387/ijdb.051994ca. Int J Dev Biol. 2005. PMID: 16096977 Review.
-
Copy number polymorphism in plant genomes.Theor Appl Genet. 2014 Jan;127(1):1-18. doi: 10.1007/s00122-013-2177-7. Epub 2013 Aug 29. Theor Appl Genet. 2014. PMID: 23989647 Free PMC article. Review.
Cited by
-
Evolutionary origin of the NCSI gene subfamily encoding norcoclaurine synthase is associated with the biosynthesis of benzylisoquinoline alkaloids in plants.Sci Rep. 2016 May 18;6:26323. doi: 10.1038/srep26323. Sci Rep. 2016. PMID: 27189519 Free PMC article.
-
A chromosome-level sequence assembly reveals the structure of the Arabidopsis thaliana Nd-1 genome and its gene set.PLoS One. 2019 May 21;14(5):e0216233. doi: 10.1371/journal.pone.0216233. eCollection 2019. PLoS One. 2019. PMID: 31112551 Free PMC article.
-
QTG-Finder2: A Generalized Machine-Learning Algorithm for Prioritizing QTL Causal Genes in Plants.G3 (Bethesda). 2020 Jul 7;10(7):2411-2421. doi: 10.1534/g3.120.401122. G3 (Bethesda). 2020. PMID: 32430305 Free PMC article.
-
Amborella gene presence/absence variation is associated with abiotic stress responses that may contribute to environmental adaptation.New Phytol. 2022 Feb;233(4):1548-1555. doi: 10.1111/nph.17658. Epub 2021 Aug 19. New Phytol. 2022. PMID: 34328223 Free PMC article. No abstract available.
-
PAV markers in Sorghum bicolour: genome pattern, affected genes and pathways, and genetic linkage map construction.Theor Appl Genet. 2015 Apr;128(4):623-37. doi: 10.1007/s00122-015-2458-4. Epub 2015 Jan 30. Theor Appl Genet. 2015. PMID: 25634103 Free PMC article.
References
-
- Morin PA, Luikart G, Wayne RK, Grp SW. SNPs in ecology, evolution and conservation. Trends Ecol Evol. 2004;19(4):208–216. doi: 10.1016/j.tree.2004.01.009. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

