Sequential evolution and escape from neutralization of simian immunodeficiency virus SIVsmE660 clones in rhesus macaques
- PMID: 22696650
- PMCID: PMC3421745
- DOI: 10.1128/JVI.00923-12
Sequential evolution and escape from neutralization of simian immunodeficiency virus SIVsmE660 clones in rhesus macaques
Abstract
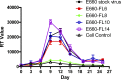
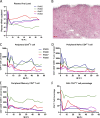
Simian immunodeficiency virus (SIV) infection of rhesus macaques has become an important surrogate model for evaluating HIV vaccine strategies. The extreme resistance to neutralizing antibody (NAb) of many commonly used strains, such as SIVmac251/239 and SIVsmE543-3, limits their potential relevance for evaluating the role of NAb in vaccine protection. In contrast, SIVsmE660 is an uncloned virus that appears to be more sensitive to neutralizing antibody. To evaluate the role of NAb in this model, we generated full-length neutralization-sensitive molecular clones of SIVsmE660 and evaluated two of these by intravenous inoculation of rhesus macaques. All animals became infected and maintained persistent viremia that was accompanied by a decline in memory CD4(+) T cells in blood and bronchoalveolar lavage fluid. High titers of autologous NAb developed by 4 weeks postinoculation but were not associated with control of viremia, and neutralization escape variants were detected concurrently with the generation of NAb. Neutralization escape was associated with substitutions and insertion/deletion polymorphisms in the V1 and V4 domains of envelope. Analysis of representative variants revealed that escape variants also induced NAbs within a few weeks of their appearance in plasma, in a pattern that is reminiscent of the escape of human immunodeficiency virus type 1 (HIV-1) isolates in humans. Although early variants maintained a neutralization-sensitive phenotype, viruses obtained later in infection were significantly less sensitive to neutralization than the parental viruses. These results indicate that NAbs exert selective pressure that drives the evolution of the SIV envelope and that this model will be useful for evaluating the role of NAb in vaccine-mediated protection.
Figures


Similar articles
-
Heterogeneity in neutralization sensitivities of viruses comprising the simian immunodeficiency virus SIVsmE660 isolate and vaccine challenge stock.J Virol. 2013 May;87(10):5477-92. doi: 10.1128/JVI.03419-12. Epub 2013 Mar 6. J Virol. 2013. PMID: 23468494 Free PMC article.
-
Breakthrough of SIV strain smE660 challenge in SIV strain mac239-vaccinated rhesus macaques despite potent autologous neutralizing antibody responses.Proc Natl Acad Sci U S A. 2015 Aug 25;112(34):10780-5. doi: 10.1073/pnas.1509731112. Epub 2015 Aug 10. Proc Natl Acad Sci U S A. 2015. PMID: 26261312 Free PMC article.
-
Correlation between env V1/V2 region diversification and neutralizing antibodies during primary infection by simian immunodeficiency virus sm in rhesus macaques.J Virol. 2004 Apr;78(7):3561-71. doi: 10.1128/jvi.78.7.3561-3571.2004. J Virol. 2004. PMID: 15016879 Free PMC article.
-
Derivation and Characterization of Pathogenic Transmitted/Founder Molecular Clones from Simian Immunodeficiency Virus SIVsmE660 and SIVmac251 following Mucosal Infection.J Virol. 2016 Sep 12;90(19):8435-53. doi: 10.1128/JVI.00718-16. Print 2016 Oct 1. J Virol. 2016. PMID: 27412591 Free PMC article.
-
Neutralization interfering antibodies: a "novel" example of humoral immune dysfunction facilitating viral escape?Viruses. 2012 Sep;4(9):1731-52. doi: 10.3390/v4091731. Epub 2012 Sep 24. Viruses. 2012. PMID: 23170181 Free PMC article. Review.
Cited by
-
Hallmarks of primate lentiviral immunodeficiency infection recapitulate loss of innate lymphoid cells.Nat Commun. 2018 Sep 27;9(1):3967. doi: 10.1038/s41467-018-05528-3. Nat Commun. 2018. PMID: 30262807 Free PMC article.
-
Simian Immunodeficiency Virus-Infected Memory CD4+ T Cells Infiltrate to the Site of Infected Macrophages in the Neuroparenchyma of a Chronic Macaque Model of Neurological Complications of AIDS.mBio. 2020 Apr 21;11(2):e00602-20. doi: 10.1128/mBio.00602-20. mBio. 2020. PMID: 32317323 Free PMC article.
-
Control of Heterologous Simian Immunodeficiency Virus SIVsmE660 Infection by DNA and Protein Coimmunization Regimens Combined with Different Toll-Like-Receptor-4-Based Adjuvants in Macaques.J Virol. 2018 Jul 17;92(15):e00281-18. doi: 10.1128/JVI.00281-18. Print 2018 Aug 1. J Virol. 2018. PMID: 29793957 Free PMC article.
-
TRIM5α Resistance Escape Mutations in the Capsid Are Transferable between Simian Immunodeficiency Virus Strains.J Virol. 2016 Nov 28;90(24):11087-11095. doi: 10.1128/JVI.01620-16. Print 2016 Dec 15. J Virol. 2016. PMID: 27681142 Free PMC article.
-
Antibody-dependent cellular cytotoxicity, infected cell binding and neutralization by antibodies to the SIV envelope glycoprotein.PLoS Pathog. 2023 May 30;19(5):e1011407. doi: 10.1371/journal.ppat.1011407. eCollection 2023 May. PLoS Pathog. 2023. PMID: 37253062 Free PMC article.
References
-
- Baba TW, et al. 2000. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat. Med. 6:200–206 - PubMed
-
- Bottiger D, Putkonen P, Oberg B. 1992. Prevention of HIV-2 and SIV infections in cynomolgus macaques by prophylactic treatment with 3′-fluorothymidine. AIDS Res. Hum. Retroviruses 8:1235–1238 - PubMed
-
- Bunnik EM, et al. 2010. Adaptation of HIV-1 envelope gp120 to humoral immunity at a population level. Nat. Med. 16:995–997 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

