Med5(Nut1) and Med17(Srb4) are direct targets of mediator histone H4 tail interactions
- PMID: 22693636
- PMCID: PMC3367926
- DOI: 10.1371/journal.pone.0038416
Med5(Nut1) and Med17(Srb4) are direct targets of mediator histone H4 tail interactions
Abstract
The Mediator complex transmits activation signals from DNA bound transcription factors to the core transcription machinery. In addition to its canonical role in transcriptional activation, recent studies have demonstrated that S. cerevisiae Mediator can interact directly with nucleosomes, and their histone tails. Mutations in Mediator subunits have shown that Mediator and certain chromatin structures mutually impact each other structurally and functionally in vivo. We have taken a UV photo cross-linking approach to further delineate the molecular basis of Mediator chromatin interactions and help determine whether the impact of certain Mediator mutants on chromatin is direct. Specifically, by using histone tail peptides substituted with an amino acid analog that is a UV activatible crosslinker, we have identified specific subunits within Mediator that participate in histone tail interactions. Using Mediator purified from mutant yeast strains we have evaluated the impact of these subunits on histone tail binding. This analysis has identified the Med5 subunit of Mediator as a target for histone tail interactions and suggests that the previously observed effect of med5 mutations on telomeric heterochromatin and silencing is direct.
Conflict of interest statement
Figures

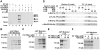


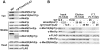


Similar articles
-
Histone modifications influence mediator interactions with chromatin.Nucleic Acids Res. 2011 Oct;39(19):8342-54. doi: 10.1093/nar/gkr551. Epub 2011 Jul 8. Nucleic Acids Res. 2011. PMID: 21742760 Free PMC article.
-
Mediator influences telomeric silencing and cellular life span.Mol Cell Biol. 2011 Jun;31(12):2413-21. doi: 10.1128/MCB.05242-11. Epub 2011 Apr 11. Mol Cell Biol. 2011. PMID: 21482672 Free PMC article.
-
The tail-module of yeast Mediator complex is required for telomere heterochromatin maintenance.Nucleic Acids Res. 2012 Jan;40(2):581-93. doi: 10.1093/nar/gkr757. Epub 2011 Sep 19. Nucleic Acids Res. 2012. PMID: 21930512 Free PMC article.
-
Med15: Glutamine-Rich Mediator Subunit with Potential for Plasticity.Trends Biochem Sci. 2019 Sep;44(9):737-751. doi: 10.1016/j.tibs.2019.03.008. Epub 2019 Apr 27. Trends Biochem Sci. 2019. PMID: 31036407 Review.
-
Acetylation of yeast histone H4 lysine 16: a switch for protein interactions in heterochromatin and euchromatin.Cold Spring Harb Symp Quant Biol. 2004;69:193-200. doi: 10.1101/sqb.2004.69.193. Cold Spring Harb Symp Quant Biol. 2004. PMID: 16117649 Review. No abstract available.
Cited by
-
Differential regulation of white-opaque switching by individual subunits of Candida albicans mediator.Eukaryot Cell. 2013 Sep;12(9):1293-304. doi: 10.1128/EC.00137-13. Epub 2013 Jul 19. Eukaryot Cell. 2013. PMID: 23873866 Free PMC article.
-
The Mediator complex and transcription regulation.Crit Rev Biochem Mol Biol. 2013 Nov-Dec;48(6):575-608. doi: 10.3109/10409238.2013.840259. Epub 2013 Oct 3. Crit Rev Biochem Mol Biol. 2013. PMID: 24088064 Free PMC article. Review.
-
Telomeric ORFs (TLOs) in Candida spp. Encode mediator subunits that regulate distinct virulence traits.PLoS Genet. 2014 Oct 30;10(10):e1004658. doi: 10.1371/journal.pgen.1004658. eCollection 2014 Oct. PLoS Genet. 2014. PMID: 25356803 Free PMC article.
-
Importance of Mediator complex in the regulation and integration of diverse signaling pathways in plants.Front Plant Sci. 2015 Sep 17;6:757. doi: 10.3389/fpls.2015.00757. eCollection 2015. Front Plant Sci. 2015. PMID: 26442070 Free PMC article. Review.
-
Transcription regulation by the Mediator complex.Nat Rev Mol Cell Biol. 2018 Apr;19(4):262-274. doi: 10.1038/nrm.2017.115. Epub 2017 Dec 6. Nat Rev Mol Cell Biol. 2018. PMID: 29209056 Review.
References
-
- Myers LC, Kornberg RD. Mediator of transcriptional regulation. Annu Rev Biochem. 2000;69:729–749. - PubMed
-
- Linder T, Gustafsson CM. The Soh1/MED31 protein is an ancient component of Schizosaccharomyces pombe and Saccharomyces cerevisiae Mediator. J Biol Chem. 2004;279:49455–49459. - PubMed
-
- Bjorklund S, Gustafsson CM. The yeast Mediator complex and its regulation. Trends Biochem Sci. 2005;30:240–244. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

