Packaging protein drugs as bacterial inclusion bodies for therapeutic applications
- PMID: 22686540
- PMCID: PMC3538617
- DOI: 10.1186/1475-2859-11-76
Packaging protein drugs as bacterial inclusion bodies for therapeutic applications
Abstract
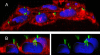
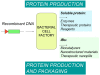
A growing number of insights on the biology of bacterial inclusion bodies (IBs) have revealed intriguing utilities of these protein particles. Since they combine mechanical stability and protein functionality, IBs have been already exploited in biocatalysis and explored for bottom-up topographical modification in tissue engineering. Being fully biocompatible and with tuneable bio-physical properties, IBs are currently emerging as agents for protein delivery into mammalian cells in protein-replacement cell therapies. So far, IBs formed by chaperones (heat shock protein 70, Hsp70), enzymes (catalase and dihydrofolate reductase), grow factors (leukemia inhibitory factor, LIF) and structural proteins (the cytoskeleton keratin 14) have been shown to rescue exposed cells from a spectrum of stresses and restore cell functions in absence of cytotoxicity. The natural penetrability of IBs into mammalian cells (reaching both cytoplasm and nucleus) empowers them as an unexpected platform for the controlled delivery of essentially any therapeutic polypeptide. Production of protein drugs by biopharma has been traditionally challenged by IB formation. However, a time might have arrived in which recombinant bacteria are to be engineered for the controlled packaging of therapeutic proteins as nanoparticulate materials (nanopills), for their extra- or intra-cellular release in medicine and cosmetics.
Figures
Similar articles
-
Functional inclusion bodies produced in bacteria as naturally occurring nanopills for advanced cell therapies.Adv Mater. 2012 Apr 3;24(13):1742-7. doi: 10.1002/adma.201104330. Epub 2012 Mar 13. Adv Mater. 2012. PMID: 22410789
-
Bacterial inclusion bodies: making gold from waste.Trends Biotechnol. 2012 Feb;30(2):65-70. doi: 10.1016/j.tibtech.2011.09.003. Epub 2011 Oct 29. Trends Biotechnol. 2012. PMID: 22037492
-
A nanostructured bacterial bioscaffold for the sustained bottom-up delivery of protein drugs.Nanomedicine (Lond). 2013 Oct;8(10):1587-99. doi: 10.2217/nnm.12.188. Epub 2013 Feb 8. Nanomedicine (Lond). 2013. PMID: 23394133
-
Bacterial Inclusion Bodies: Discovering Their Better Half.Trends Biochem Sci. 2017 Sep;42(9):726-737. doi: 10.1016/j.tibs.2017.01.005. Epub 2017 Feb 27. Trends Biochem Sci. 2017. PMID: 28254353 Review.
-
Bacterial inclusion bodies are industrially exploitable amyloids.FEMS Microbiol Rev. 2019 Jan 1;43(1):53-72. doi: 10.1093/femsre/fuy038. FEMS Microbiol Rev. 2019. PMID: 30357330 Review.
Cited by
-
Active inclusion bodies of acid phosphatase PhoC: aggregation induced by GFP fusion and activities modulated by linker flexibility.Microb Cell Fact. 2013 Mar 14;12:25. doi: 10.1186/1475-2859-12-25. Microb Cell Fact. 2013. PMID: 23497261 Free PMC article.
-
Trends in recombinant protein use in animal production.Microb Cell Fact. 2017 Mar 4;16(1):40. doi: 10.1186/s12934-017-0654-4. Microb Cell Fact. 2017. PMID: 28259156 Free PMC article. Review.
-
Bacterial mimetics of endocrine secretory granules as immobilized in vivo depots for functional protein drugs.Sci Rep. 2016 Oct 24;6:35765. doi: 10.1038/srep35765. Sci Rep. 2016. PMID: 27775083 Free PMC article.
-
Bacterial inclusion bodies as potential synthetic devices for pathogen recognition and a therapeutic substance release.Microb Cell Fact. 2013 Feb 7;12:16. doi: 10.1186/1475-2859-12-16. Microb Cell Fact. 2013. PMID: 23391325 Free PMC article.
-
The Poly-Histidine Tag H6 Mediates Structural and Functional Properties of Disintegrating, Protein-Releasing Inclusion Bodies.Pharmaceutics. 2022 Mar 10;14(3):602. doi: 10.3390/pharmaceutics14030602. Pharmaceutics. 2022. PMID: 35335976 Free PMC article.
References
-
- Jevsevar S, Gaberc-Porekar V, Fonda I, Podobnik B, Grdadolnik J, Menart V. Production of nonclassical inclusion bodies from which correctly folded protein can be extracted. Biotechnol Prog. 2005;21:632–639. - PubMed
-
- Garcia-Fruitos E, Vazquez E, Diez-Gil C, Corchero JL, Seras-Franzoso J, Ratera I. et al.Bacterial inclusion bodies: making gold from waste. Trends Biotechnol. 2012;30:65–70. - PubMed
-
- Garcia-Fruitos E, Villaverde A. Friendly production of bacterial inclusion bodies. Korean J Chem Eng. 2010;27:385–389.
-
- Nahalka J, Nidetzky B. Fusion to a pull-down domain: a novel approach of producingTrigonopsis variabilisD-amino acid oxidase as insoluble enzyme aggregates. Biotechnol Bioeng. 2007;97:454–461. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

