Molecular dynamics and mutational analysis of the catalytic and translocation cycle of RNA polymerase
- PMID: 22676913
- PMCID: PMC3533926
- DOI: 10.1186/2046-1682-5-11
Molecular dynamics and mutational analysis of the catalytic and translocation cycle of RNA polymerase
Abstract
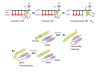
Background: During elongation, multi-subunit RNA polymerases (RNAPs) cycle between phosphodiester bond formation and nucleic acid translocation. In the conformation associated with catalysis, the mobile "trigger loop" of the catalytic subunit closes on the nucleoside triphosphate (NTP) substrate. Closing of the trigger loop is expected to exclude water from the active site, and dehydration may contribute to catalysis and fidelity. In the absence of a NTP substrate in the active site, the trigger loop opens, which may enable translocation. Another notable structural element of the RNAP catalytic center is the "bridge helix" that separates the active site from downstream DNA. The bridge helix may participate in translocation by bending against the RNA/DNA hybrid to induce RNAP forward movement and to vacate the active site for the next NTP loading. The transition between catalytic and translocation conformations of RNAP is not evident from static crystallographic snapshots in which macromolecular motions may be restrained by crystal packing.
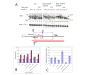
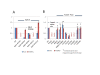
Results: All atom molecular dynamics simulations of Thermus thermophilus (Tt) RNAP reveal flexible hinges, located within the two helices at the base of the trigger loop, and two glycine hinges clustered near the N-terminal end of the bridge helix. As simulation progresses, these hinges adopt distinct conformations in the closed and open trigger loop structures. A number of residues (described as "switch" residues) trade atomic contacts (ion pairs or hydrogen bonds) in response to changes in hinge orientation. In vivo phenotypes and in vitro activities rendered by mutations in the hinge and switch residues in Saccharomyces cerevisiae (Sc) RNAP II support the importance of conformational changes predicted from simulations in catalysis and translocation. During simulation, the elongation complex with an open trigger loop spontaneously translocates forward relative to the elongation complex with a closed trigger loop.
Conclusions: Switching between catalytic and translocating RNAP forms involves closing and opening of the trigger loop and long-range conformational changes in the atomic contacts of amino acid side chains, some located at a considerable distance from the trigger loop and active site. Trigger loop closing appears to support chemistry and the fidelity of RNA synthesis. Trigger loop opening and limited bridge helix bending appears to promote forward nucleic acid translocation.
Figures






Similar articles
-
Conformational coupling, bridge helix dynamics and active site dehydration in catalysis by RNA polymerase.Biochim Biophys Acta. 2010 Aug;1799(8):575-87. doi: 10.1016/j.bbagrm.2010.05.002. Epub 2010 May 15. Biochim Biophys Acta. 2010. PMID: 20478425 Free PMC article.
-
Hinge action versus grip in translocation by RNA polymerase.Transcription. 2018;9(1):1-16. doi: 10.1080/21541264.2017.1330179. Epub 2017 Aug 30. Transcription. 2018. PMID: 28853995 Free PMC article.
-
The RNA polymerase bridge helix YFI motif in catalysis, fidelity and translocation.Biochim Biophys Acta. 2013 Feb;1829(2):187-98. doi: 10.1016/j.bbagrm.2012.11.005. Epub 2012 Nov 30. Biochim Biophys Acta. 2013. PMID: 23202476 Free PMC article.
-
NTP-driven translocation and regulation of downstream template opening by multi-subunit RNA polymerases.Biochem Cell Biol. 2005 Aug;83(4):486-96. doi: 10.1139/o05-059. Biochem Cell Biol. 2005. PMID: 16094452 Review.
-
Enzyme dynamics from NMR spectroscopy.Acc Chem Res. 2015 Feb 17;48(2):457-65. doi: 10.1021/ar500340a. Epub 2015 Jan 9. Acc Chem Res. 2015. PMID: 25574774 Free PMC article. Review.
Cited by
-
Active Center Control of Termination by RNA Polymerase III and tRNA Gene Transcription Levels In Vivo.PLoS Genet. 2016 Aug 12;12(8):e1006253. doi: 10.1371/journal.pgen.1006253. eCollection 2016 Aug. PLoS Genet. 2016. PMID: 27518095 Free PMC article.
-
Computational simulation strategies for analysis of multisubunit RNA polymerases.Chem Rev. 2013 Nov 13;113(11):8546-66. doi: 10.1021/cr400046x. Epub 2013 Aug 29. Chem Rev. 2013. PMID: 23987500 Free PMC article. Review. No abstract available.
-
Divergent contributions of conserved active site residues to transcription by eukaryotic RNA polymerases I and II.Cell Rep. 2013 Sep 12;4(5):974-84. doi: 10.1016/j.celrep.2013.07.044. Epub 2013 Aug 29. Cell Rep. 2013. PMID: 23994471 Free PMC article.
-
Trigger-helix folding pathway and SI3 mediate catalysis and hairpin-stabilized pausing by Escherichia coli RNA polymerase.Nucleic Acids Res. 2014 Nov 10;42(20):12707-21. doi: 10.1093/nar/gku997. Epub 2014 Oct 21. Nucleic Acids Res. 2014. PMID: 25336618 Free PMC article.
-
Millisecond dynamics of RNA polymerase II translocation at atomic resolution.Proc Natl Acad Sci U S A. 2014 May 27;111(21):7665-70. doi: 10.1073/pnas.1315751111. Epub 2014 Apr 21. Proc Natl Acad Sci U S A. 2014. PMID: 24753580 Free PMC article.
References
-
- Kireeva M, Kashlev M, Burton ZF. Translocation by multi-subunit RNA polymerases. Biochim Biophys Acta. 2010;1799:389–401. - PubMed
-
- Nedialkov YA, Gong XQ, Hovde SL, Yamaguchi Y, Handa H, Geiger JH, Yan H, Burton ZF. NTP-driven translocation by human RNA polymerase II. J Biol Chem. 2003;278(20):18303–18312. - PubMed
-
- Xiong Y, Burton ZF. A Tunable Ratchet Driving Human RNA Polymerase II Translocation Adjusted by Accurately Templated Nucleoside Triphosphates Loaded at Downstream Sites and by Elongation Factors. J Biol Chem. 2007;282(50):36582–36592. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

