TGR5 potentiates GLP-1 secretion in response to anionic exchange resins
- PMID: 22666533
- PMCID: PMC3362799
- DOI: 10.1038/srep00430
TGR5 potentiates GLP-1 secretion in response to anionic exchange resins
Abstract
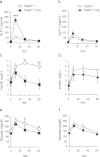
Anionic exchange resins are bona fide cholesterol-lowering agents with glycemia lowering actions in diabetic patients. Potentiation of intestinal GLP-1 secretion has been proposed to contribute to the glycemia lowering effect of these non-systemic drugs. Here, we show that resin exposure enhances GLP-1 secretion and improves glycemic control in diet-induced animal models of "diabesity", effects which are critically dependent on TGR5, a G protein-coupled receptor that is activated by bile acids. We identified the colon as a major source of GLP-1 secretion after resin treatment. Furthermore, we demonstrate that the boost in GLP-1 release by resins is due to both enhanced TGR5-dependent production of the precursor transcript of GLP-1 as well as to the local enrichment of TGR5 agonists in the colon. Thus, TGR5 represents an essential component in the pathway mediating the enhanced GLP-1 release in response to anionic exchange resins.
Figures

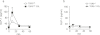
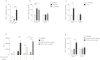
Similar articles
-
Farnesoid X receptor inhibits glucagon-like peptide-1 production by enteroendocrine L cells.Nat Commun. 2015 Jul 2;6:7629. doi: 10.1038/ncomms8629. Nat Commun. 2015. PMID: 26134028 Free PMC article.
-
TGR5-mediated bile acid sensing controls glucose homeostasis.Cell Metab. 2009 Sep;10(3):167-77. doi: 10.1016/j.cmet.2009.08.001. Cell Metab. 2009. PMID: 19723493 Free PMC article.
-
Activation of Transmembrane Bile Acid Receptor TGR5 Modulates Pancreatic Islet α Cells to Promote Glucose Homeostasis.J Biol Chem. 2016 Mar 25;291(13):6626-40. doi: 10.1074/jbc.M115.699504. Epub 2016 Jan 12. J Biol Chem. 2016. PMID: 26757816 Free PMC article.
-
Bile-induced secretion of glucagon-like peptide-1: pathophysiological implications in type 2 diabetes?Am J Physiol Endocrinol Metab. 2010 Jul;299(1):E10-3. doi: 10.1152/ajpendo.00137.2010. Epub 2010 Apr 27. Am J Physiol Endocrinol Metab. 2010. PMID: 20424139 Review.
-
Physiology of proglucagon peptides: role of glucagon and GLP-1 in health and disease.Physiol Rev. 2015 Apr;95(2):513-48. doi: 10.1152/physrev.00013.2014. Physiol Rev. 2015. PMID: 25834231 Review.
Cited by
-
Design, synthesis and evaluation of 3-phenoxypyrazine-2-carboxamide derivatives as potent TGR5 agonists.RSC Adv. 2022 Jan 27;12(6):3618-3629. doi: 10.1039/d1ra08867j. eCollection 2022 Jan 24. RSC Adv. 2022. PMID: 35425398 Free PMC article.
-
Biological tuners to reshape the bile acid pool for therapeutic purposes in non-alcoholic fatty liver disease.Clin Sci (Lond). 2023 Jan 13;137(1):65-85. doi: 10.1042/CS20220697. Clin Sci (Lond). 2023. PMID: 36601783 Free PMC article. Review.
-
Colesevelam attenuates cholestatic liver and bile duct injury in Mdr2-/- mice by modulating composition, signalling and excretion of faecal bile acids.Gut. 2018 Sep;67(9):1683-1691. doi: 10.1136/gutjnl-2017-314553. Epub 2018 Apr 10. Gut. 2018. PMID: 29636383 Free PMC article.
-
Bariatric surgery reveals a gut-restricted TGR5 agonist with anti-diabetic effects.Nat Chem Biol. 2021 Jan;17(1):20-29. doi: 10.1038/s41589-020-0604-z. Epub 2020 Aug 3. Nat Chem Biol. 2021. PMID: 32747812 Free PMC article.
-
The bile acid TGR5 membrane receptor: from basic research to clinical application.Dig Liver Dis. 2014 Apr;46(4):302-12. doi: 10.1016/j.dld.2013.10.021. Epub 2014 Jan 9. Dig Liver Dis. 2014. PMID: 24411485 Free PMC article. Review.
References
-
- Thomas C., Pellicciari R., Pruzanski M., Auwerx J. & Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov 7, 678–693 (2008). - PubMed
-
- Trauner M., Claudel T., Fickert P., Moustafa T. & Wagner M. Bile acids as regulators of hepatic lipid and glucose metabolism. Dig Dis 28, 220–224 (2010). - PubMed
-
- Kawamata Y. et al. A G protein-coupled receptor responsive to bile acids. J Biol Chem 278, 9435–9440 (2003). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

