Identification of novel subgroup A variants with enhanced receptor binding and replicative capacity in primary isolates of anaemogenic strains of feline leukaemia virus
- PMID: 22650160
- PMCID: PMC3403869
- DOI: 10.1186/1742-4690-9-48
Identification of novel subgroup A variants with enhanced receptor binding and replicative capacity in primary isolates of anaemogenic strains of feline leukaemia virus
Abstract
Background: The development of anaemia in feline leukaemia virus (FeLV)-infected cats is associated with the emergence of a novel viral subgroup, FeLV-C. FeLV-C arises from the subgroup that is transmitted, FeLV-A, through alterations in the amino acid sequence of the receptor binding domain (RBD) of the envelope glycoprotein that result in a shift in the receptor usage and the cell tropism of the virus. The factors that influence the transition from subgroup A to subgroup C remain unclear, one possibility is that a selective pressure in the host drives the acquisition of mutations in the RBD, creating A/C intermediates with enhanced abilities to interact with the FeLV-C receptor, FLVCR. In order to understand further the emergence of FeLV-C in the infected cat, we examined primary isolates of FeLV-C for evidence of FeLV-A variants that bore mutations consistent with a gradual evolution from FeLV-A to FeLV-C.
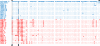
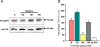
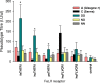
Results: Within each isolate of FeLV-C, we identified variants that were ostensibly subgroup A by nucleic acid sequence comparisons, but which bore mutations in the RBD. One such mutation, N91D, was present in multiple isolates and when engineered into a molecular clone of the prototypic FeLV-A (Glasgow-1), enhanced replication was noted in feline cells. Expression of the N91D Env on murine leukaemia virus (MLV) pseudotypes enhanced viral entry mediated by the FeLV-A receptor THTR1 while soluble FeLV-A Env bearing the N91D mutation bound more efficiently to mouse or guinea pig cells bearing the FeLV-A and -C receptors. Long-term in vitro culture of variants bearing the N91D substitution in the presence of anti-FeLV gp70 antibodies did not result in the emergence of FeLV-C variants, suggesting that additional selective pressures in the infected cat may drive the subsequent evolution from subgroup A to subgroup C.
Conclusions: Our data support a model in which variants of FeLV-A, bearing subtle differences in the RBD of Env, may be predisposed towards enhanced replication in vivo and subsequent conversion to FeLV-C. The selection pressures in vivo that drive the emergence of FeLV-C in a proportion of infected cats remain to be established.
Figures
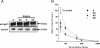




Similar articles
-
Identification of a feline leukemia virus variant that can use THTR1, FLVCR1, and FLVCR2 for infection.J Virol. 2009 Jul;83(13):6706-16. doi: 10.1128/JVI.02317-08. Epub 2009 Apr 15. J Virol. 2009. PMID: 19369334 Free PMC article.
-
Reduced Folate Carrier: an Entry Receptor for a Novel Feline Leukemia Virus Variant.J Virol. 2019 Jun 14;93(13):e00269-19. doi: 10.1128/JVI.00269-19. Print 2019 Jul 1. J Virol. 2019. PMID: 30996094 Free PMC article.
-
Partial dissociation of subgroup C phenotype and in vivo behaviour in feline leukaemia viruses with chimeric envelope genes.J Gen Virol. 1992 Nov;73 ( Pt 11):2839-47. doi: 10.1099/0022-1317-73-11-2839. J Gen Virol. 1992. PMID: 1331290
-
Endogenous env elements: partners in generation of pathogenic feline leukemia viruses.Virus Genes. 1995;11(2-3):147-61. doi: 10.1007/BF01728655. Virus Genes. 1995. PMID: 8828142 Review.
-
Control of feline leukaemia virus.Vet Immunol Immunopathol. 1989 May;21(1):69-83. doi: 10.1016/0165-2427(89)90131-1. Vet Immunol Immunopathol. 1989. PMID: 2549695 Review.
Cited by
-
On the concept and elucidation of endogenous retroviruses.Philos Trans R Soc Lond B Biol Sci. 2013 Aug 12;368(1626):20120494. doi: 10.1098/rstb.2012.0494. Print 2013 Sep 19. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 23938748 Free PMC article. Review.
-
Prevalence and Genomic Diversity of Feline Leukemia Virus in Privately Owned and Shelter Cats in Aburrá Valley, Colombia.Viruses. 2020 Apr 20;12(4):464. doi: 10.3390/v12040464. Viruses. 2020. PMID: 32325926 Free PMC article.
-
Novel Feline Leukemia Virus Interference Group Based on the env Gene.J Virol. 2016 Apr 14;90(9):4832-4837. doi: 10.1128/JVI.03229-15. Print 2016 May. J Virol. 2016. PMID: 26889025 Free PMC article.
-
Feline Leukemia Virus Frequently Spills Over from Domestic Cats to North American Pumas.J Virol. 2022 Dec 14;96(23):e0120122. doi: 10.1128/jvi.01201-22. Epub 2022 Nov 14. J Virol. 2022. PMID: 36374109 Free PMC article.
References
-
- Hardy WD, Hess PW, MacEwen EG, McClelland AJ, Zuckerman EE, Essex M, Cotter SM, Jarrett O. Biology of feline leukemia virus in the natural environment. Cancer Res. 1976;36:582–588. - PubMed
-
- Hardy WD, Hirshaut Y, Hess P. Detection of the feline leukemia virus and other mammalian oncornaviruses by immunofluorescence. Bibl Haematol. 1973;39:778–799. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

