The role of translation initiation regulation in haematopoiesis
- PMID: 22649283
- PMCID: PMC3357504
- DOI: 10.1155/2012/576540
The role of translation initiation regulation in haematopoiesis
Abstract
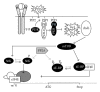
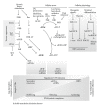
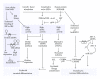
Organisation of RNAs into functional subgroups that are translated in response to extrinsic and intrinsic factors underlines a relatively unexplored gene expression modulation that drives cell fate in the same manner as regulation of the transcriptome by transcription factors. Recent studies on the molecular mechanisms of inflammatory responses and haematological disorders indicate clearly that the regulation of mRNA translation at the level of translation initiation, mRNA stability, and protein isoform synthesis is implicated in the tight regulation of gene expression. This paper outlines how these posttranscriptional control mechanisms, including control at the level of translation initiation factors and the role of RNA binding proteins, affect hematopoiesis. The clinical relevance of these mechanisms in haematological disorders indicates clearly the potential therapeutic implications and the need of molecular tools that allow measurement at the level of translational control. Although the importance of miRNAs in translation control is well recognised and studied extensively, this paper will exclude detailed account of this level of control.
Figures
Similar articles
-
The RNA Helicase DeaD Stimulates ExsA Translation To Promote Expression of the Pseudomonas aeruginosa Type III Secretion System.J Bacteriol. 2015 Aug;197(16):2664-74. doi: 10.1128/JB.00231-15. Epub 2015 Jun 8. J Bacteriol. 2015. PMID: 26055113 Free PMC article.
-
CsrA-Mediated Translational Activation of ymdA Expression in Escherichia coli.mBio. 2020 Sep 15;11(5):e00849-20. doi: 10.1128/mBio.00849-20. mBio. 2020. PMID: 32934077 Free PMC article.
-
Cell Fate Control by Translation: mRNA Translation Initiation as a Therapeutic Target for Cancer Development and Stem Cell Fate Control.Biomolecules. 2019 Oct 29;9(11):665. doi: 10.3390/biom9110665. Biomolecules. 2019. PMID: 31671902 Free PMC article. Review.
-
Engineering bacterial translation initiation - Do we have all the tools we need?Biochim Biophys Acta Gen Subj. 2017 Nov;1861(11 Pt B):3060-3069. doi: 10.1016/j.bbagen.2017.03.008. Epub 2017 Mar 14. Biochim Biophys Acta Gen Subj. 2017. PMID: 28315412 Review.
-
Ribosome profiling reveals translation control as a key mechanism generating differential gene expression in Trypanosoma cruzi.BMC Genomics. 2015 Jun 9;16(1):443. doi: 10.1186/s12864-015-1563-8. BMC Genomics. 2015. PMID: 26054634 Free PMC article.
Cited by
-
Expression of different functional isoforms in haematopoiesis.Int J Hematol. 2014 Jan;99(1):4-11. doi: 10.1007/s12185-013-1477-7. Epub 2013 Dec 1. Int J Hematol. 2014. PMID: 24293279 Review.
-
Rapamycin treatment dose-dependently improves the cystic kidney in a new ADPKD mouse model via the mTORC1 and cell-cycle-associated CDK1/cyclin axis.J Cell Mol Med. 2017 Aug;21(8):1619-1635. doi: 10.1111/jcmm.13091. Epub 2017 Feb 28. J Cell Mol Med. 2017. PMID: 28244683 Free PMC article.
-
Multifaced regulator: RNA binding proteins and their roles in hematopoiesis.Blood Sci. 2019 Sep 17;1(1):69-72. doi: 10.1097/BS9.0000000000000008. eCollection 2019 Aug. Blood Sci. 2019. PMID: 35402803 Free PMC article.
-
Evidence for a functionally relevant rocaglamide binding site on the eIF4A-RNA complex.ACS Chem Biol. 2013 Jul 19;8(7):1519-27. doi: 10.1021/cb400158t. Epub 2013 May 7. ACS Chem Biol. 2013. PMID: 23614532 Free PMC article.
-
Mutation of Gemin5 Causes Defective Hematopoietic Stem/Progenitor Cells Proliferation in Zebrafish Embryonic Hematopoiesis.Front Cell Dev Biol. 2021 Apr 30;9:670654. doi: 10.3389/fcell.2021.670654. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33996826 Free PMC article.
References
-
- Phillips RL, Ernst RE, Brunk B, et al. The genetic program of hematopoietic stem cells. Science. 2000;288(5471):1635–1640. - PubMed
-
- Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265(5178):1573–1577. - PubMed
-
- Gingras AC, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes and Development. 2001;15(7):807–826. - PubMed
LinkOut - more resources
Full Text Sources

