Evolution of saxitoxin synthesis in cyanobacteria and dinoflagellates
- PMID: 22628533
- PMCID: PMC3525144
- DOI: 10.1093/molbev/mss142
Evolution of saxitoxin synthesis in cyanobacteria and dinoflagellates
Abstract
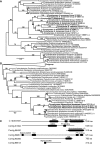
Dinoflagellates produce a variety of toxic secondary metabolites that have a significant impact on marine ecosystems and fisheries. Saxitoxin (STX), the cause of paralytic shellfish poisoning, is produced by three marine dinoflagellate genera and is also made by some freshwater cyanobacteria. Genes involved in STX synthesis have been identified in cyanobacteria but are yet to be reported in the massive genomes of dinoflagellates. We have assembled comprehensive transcriptome data sets for several STX-producing dinoflagellates and a related non-toxic species and have identified 265 putative homologs of 13 cyanobacterial STX synthesis genes, including all of the genes directly involved in toxin synthesis. Putative homologs of four proteins group closely in phylogenies with cyanobacteria and are likely the functional homologs of sxtA, sxtG, and sxtB in dinoflagellates. However, the phylogenies do not support the transfer of these genes directly between toxic cyanobacteria and dinoflagellates. SxtA is split into two proteins in the dinoflagellates corresponding to the N-terminal portion containing the methyltransferase and acyl carrier protein domains and a C-terminal portion with the aminotransferase domain. Homologs of sxtB and N-terminal sxtA are present in non-toxic strains, suggesting their functions may not be limited to saxitoxin production. Only homologs of the C-terminus of sxtA and sxtG were found exclusively in toxic strains. A more thorough survey of STX+ dinoflagellates will be needed to determine if these two genes may be specific to SXT production in dinoflagellates. The A. tamarense transcriptome does not contain homologs for the remaining STX genes. Nevertheless, we identified candidate genes with similar predicted biochemical activities that account for the missing functions. These results suggest that the STX synthesis pathway was likely assembled independently in the distantly related cyanobacteria and dinoflagellates, although using some evolutionarily related proteins. The biological role of STX is not well understood in either cyanobacteria or dinoflagellates. However, STX production in these two ecologically distinct groups of organisms suggests that this toxin confers a benefit to producers that we do not yet fully understand.
Figures

Similar articles
-
Gene duplication, loss and selection in the evolution of saxitoxin biosynthesis in alveolates.Mol Phylogenet Evol. 2015 Nov;92:165-80. doi: 10.1016/j.ympev.2015.06.017. Epub 2015 Jun 30. Mol Phylogenet Evol. 2015. PMID: 26140862
-
Transcriptome survey and toxin measurements reveal evolutionary modification and loss of saxitoxin biosynthesis genes in the dinoflagellates Amphidinium carterae and Prorocentrum micans.Ecotoxicol Environ Saf. 2020 Jun 1;195:110474. doi: 10.1016/j.ecoenv.2020.110474. Epub 2020 Mar 19. Ecotoxicol Environ Saf. 2020. PMID: 32200147
-
Polyphyletic origin of saxitoxin biosynthesis genes in the marine dinoflagellate Alexandrium revealed by comparative transcriptomics.Harmful Algae. 2024 Apr;134:102620. doi: 10.1016/j.hal.2024.102620. Epub 2024 Mar 20. Harmful Algae. 2024. PMID: 38705616
-
Evolution and distribution of saxitoxin biosynthesis in dinoflagellates.Mar Drugs. 2013 Aug 8;11(8):2814-28. doi: 10.3390/md11082814. Mar Drugs. 2013. PMID: 23966031 Free PMC article. Review.
-
Biosynthesis of Saxitoxin in Marine Dinoflagellates: An Omics Perspective.Mar Drugs. 2020 Feb 5;18(2):103. doi: 10.3390/md18020103. Mar Drugs. 2020. PMID: 32033403 Free PMC article. Review.
Cited by
-
When naked became armored: an eight-gene phylogeny reveals monophyletic origin of theca in dinoflagellates.PLoS One. 2012;7(11):e50004. doi: 10.1371/journal.pone.0050004. Epub 2012 Nov 19. PLoS One. 2012. PMID: 23185516 Free PMC article.
-
Exposure to bloom-like concentrations of two marine Synechococcus cyanobacteria (strains CC9311 and CC9902) differentially alters fish behaviour.Conserv Physiol. 2014 Jun 5;2(1):cou020. doi: 10.1093/conphys/cou020. eCollection 2014. Conserv Physiol. 2014. PMID: 27293641 Free PMC article.
-
The Genetic Basis of Toxin Biosynthesis in Dinoflagellates.Microorganisms. 2019 Jul 29;7(8):222. doi: 10.3390/microorganisms7080222. Microorganisms. 2019. PMID: 31362398 Free PMC article. Review.
-
RNA Sequencing Revealed Numerous Polyketide Synthase Genes in the Harmful Dinoflagellate Karenia mikimotoi.PLoS One. 2015 Nov 11;10(11):e0142731. doi: 10.1371/journal.pone.0142731. eCollection 2015. PLoS One. 2015. PMID: 26561394 Free PMC article.
-
Bacterial community affects toxin production by Gymnodinium catenatum.PLoS One. 2014 Aug 12;9(8):e104623. doi: 10.1371/journal.pone.0104623. eCollection 2014. PLoS One. 2014. PMID: 25117053 Free PMC article.
References
-
- Anderson DM, Kulis DM, Doucette GJ, Gallager JC, Balech E. Biogeography of toxic dinoflagellates in the genus Alexandrium from the northeast United States and Canada as determined by morphology, bioluminescence, toxin composition, and mating compatibility. Mar Biol. 1994;120:467–478.
-
- Birol I, Jackman SD, Nielsen CB, et al. (15 co-authors) De novo transcriptome assembly with ABySS. Bioinformatics. 2009;25:2872–2877. - PubMed
-
- Bowler C, Allen AE, Badger JH, et al. (77 co-authors) The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–244. - PubMed
-
- Brosnahan ML, Kulis DM, Solow AR, Erdner DL, Percy L, Lewis J, Anderson DM. Outbreeding lethality between toxic Group I and nontoxic Group III Alexandrium tamarense spp. isolates: predominance of heterotypic encystment and implications for mating interactions and biogeography. Deep-Sea Res. II. 2010;57(3–4):175–189. - PMC - PubMed
-
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

