Thioredoxin and thioredoxin target proteins: from molecular mechanisms to functional significance
- PMID: 22607099
- PMCID: PMC3579385
- DOI: 10.1089/ars.2011.4322
Thioredoxin and thioredoxin target proteins: from molecular mechanisms to functional significance
Abstract
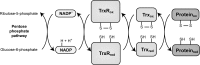
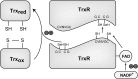
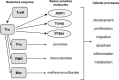
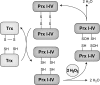
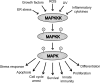
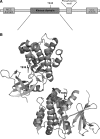
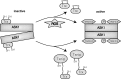
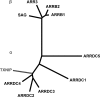
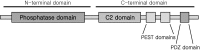
The thioredoxin (Trx) system is one of the central antioxidant systems in mammalian cells, maintaining a reducing environment by catalyzing electron flux from nicotinamide adenine dinucleotide phosphate through Trx reductase to Trx, which reduces its target proteins using highly conserved thiol groups. While the importance of protecting cells from the detrimental effects of reactive oxygen species is clear, decades of research in this field revealed that there is a network of redox-sensitive proteins forming redox-dependent signaling pathways that are crucial for fundamental cellular processes, including metabolism, proliferation, differentiation, migration, and apoptosis. Trx participates in signaling pathways interacting with different proteins to control their dynamic regulation of structure and function. In this review, we focus on Trx target proteins that are involved in redox-dependent signaling pathways. Specifically, Trx-dependent reductive enzymes that participate in classical redox reactions and redox-sensitive signaling molecules are discussed in greater detail. The latter are extensively discussed, as ongoing research unveils more and more details about the complex signaling networks of Trx-sensitive signaling molecules such as apoptosis signal-regulating kinase 1, Trx interacting protein, and phosphatase and tensin homolog, thus highlighting the potential direct and indirect impact of their redox-dependent interaction with Trx. Overall, the findings that are described here illustrate the importance and complexity of Trx-dependent, redox-sensitive signaling in the cell. Our increasing understanding of the components and mechanisms of these signaling pathways could lead to the identification of new potential targets for the treatment of diseases, including cancer and diabetes.
Figures


Similar articles
-
The thioredoxin antioxidant system.Free Radic Biol Med. 2014 Jan;66:75-87. doi: 10.1016/j.freeradbiomed.2013.07.036. Epub 2013 Jul 27. Free Radic Biol Med. 2014. PMID: 23899494 Review.
-
Redox Signaling Mediated by Thioredoxin and Glutathione Systems in the Central Nervous System.Antioxid Redox Signal. 2017 Nov 1;27(13):989-1010. doi: 10.1089/ars.2016.6925. Epub 2017 May 18. Antioxid Redox Signal. 2017. PMID: 28443683 Free PMC article. Review.
-
Thioredoxin and redox signaling: Roles of the thioredoxin system in control of cell fate.Arch Biochem Biophys. 2017 Mar 1;617:101-105. doi: 10.1016/j.abb.2016.09.011. Epub 2016 Sep 22. Arch Biochem Biophys. 2017. PMID: 27665998 Review.
-
Thioredoxin Signaling Pathways in Cancer.Antioxid Redox Signal. 2023 Feb;38(4-6):403-424. doi: 10.1089/ars.2022.0074. Epub 2022 Aug 9. Antioxid Redox Signal. 2023. PMID: 35686449 Review.
-
Thioredoxin system in cell death progression.Antioxid Redox Signal. 2012 Dec 15;17(12):1738-47. doi: 10.1089/ars.2012.4650. Epub 2012 Jun 11. Antioxid Redox Signal. 2012. PMID: 22530689 Review.
Cited by
-
Thioredoxin-interacting protein mediates high glucose-induced reactive oxygen species generation by mitochondria and the NADPH oxidase, Nox4, in mesangial cells.J Biol Chem. 2013 Mar 8;288(10):6835-48. doi: 10.1074/jbc.M112.419101. Epub 2013 Jan 17. J Biol Chem. 2013. PMID: 23329835 Free PMC article.
-
The Emerging Role of Thioredoxin-Interacting Protein in Myocardial Ischemia/Reperfusion Injury.J Cardiovasc Pharmacol Ther. 2017 May;22(3):219-229. doi: 10.1177/1074248416675731. Epub 2016 Nov 2. J Cardiovasc Pharmacol Ther. 2017. PMID: 27807222 Free PMC article. Review.
-
Dual inhibition of thioredoxin reductase and proteasome is required for auranofin-induced paraptosis in breast cancer cells.Cell Death Dis. 2023 Jan 19;14(1):42. doi: 10.1038/s41419-023-05586-6. Cell Death Dis. 2023. PMID: 36658130 Free PMC article.
-
Thioredoxin 1 and Thioredoxin Reductase 1 Redox System Is Dysregulated in Neutrophils of Subjects with Autism: In Vitro Effects of Environmental Toxicant, Methylmercury.Toxics. 2023 Aug 29;11(9):739. doi: 10.3390/toxics11090739. Toxics. 2023. PMID: 37755749 Free PMC article.
-
DciA Helicase Operators Exhibit Diversity across Bacterial Phyla.J Bacteriol. 2022 Aug 16;204(8):e0016322. doi: 10.1128/jb.00163-22. Epub 2022 Jul 26. J Bacteriol. 2022. PMID: 35880876 Free PMC article.
References
-
- Ago T. Liu T. Zhai P. Chen W. Li H. Molkentin JD. Vatner SF. Sadoshima J. A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy. Cell. 2008;133:978–993. - PubMed
-
- Ahsan MK. Masutani H. Yamaguchi Y. Kim Y-C. Nosaka K. Matsuoka M. Nishinaka Y. Maeda M. Yodoi J. Loss of interleukin-2-dependency in HTLV-I-infected T cells on gene silencing of thioredoxin-binding protein-2. Oncogene. 2005;25:2181–2191. - PubMed
-
- Ahsan MK. Okuyama H. Hoshino Y. Oka S-I. Masutani H. Yodoi J. Nakamura H. Thioredoxin-binding protein-2 deficiency enhances methionine-choline deficient diet-induced hepatic steatosis but inhibits steatohepatitis in mice. Antioxid Redox Signal. 2009;11:2573–2584. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

