PIDDosome-independent tumor suppression by Caspase-2
- PMID: 22595758
- PMCID: PMC3438502
- DOI: 10.1038/cdd.2012.54
PIDDosome-independent tumor suppression by Caspase-2
Abstract
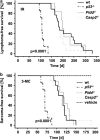
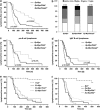
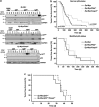
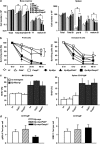
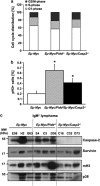
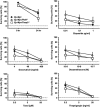
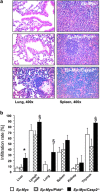
The PIDDosome, a multiprotein complex constituted of the 'p53-induced protein with a death domain (PIDD), 'receptor-interacting protein (RIP)-associated ICH-1/CED-3 homologous protein with a death domain' (RAIDD) and pro-Caspase-2 has been defined as an activating platform for this apoptosis-related protease. PIDD has been implicated in p53-mediated cell death in response to DNA damage but also in DNA repair and nuclear factor kappa-light-chain enhancer (NF-κB) activation upon genotoxic stress, together with RIP-1 kinase and Nemo/IKKγ. As all these cellular responses are critical for tumor suppression and deregulated expression of individual PIDDosome components has been noted in human cancer, we investigated their role in oncogenesis induced by DNA damage or oncogenic stress in gene-ablated mice. We observed that Pidd or Caspase-2 failed to suppress lymphoma formation triggered by γ-irradiation or 3-methylcholanthrene-driven fibrosarcoma development. In contrast, Caspase-2 showed tumor suppressive capacity in response to aberrant c-Myc expression, which did not rely on PIDD, the BH3-only protein Bid (BH3 interacting domain death agonist) or the death receptor ligand Trail (TNF-related apoptosis-inducing ligand), but associated with reduced rates of p53 loss and increased extranodal dissemination of tumor cells. In contrast, Pidd deficiency associated with abnormal M-phase progression and delayed disease onset, indicating that both proteins are differentially engaged upon oncogenic stress triggered by c-Myc, leading to opposing effects on tumor-free survival.
Figures
Similar articles
-
The tumor-modulatory effects of Caspase-2 and Pidd1 do not require the scaffold protein Raidd.Cell Death Differ. 2015 Nov;22(11):1803-11. doi: 10.1038/cdd.2015.31. Epub 2015 Apr 10. Cell Death Differ. 2015. PMID: 25857265 Free PMC article.
-
Molecular basis of neurodevelopmental disorders caused by pathogenic variants of PIDD.Biochem Biophys Res Commun. 2023 Feb 19;645:147-153. doi: 10.1016/j.bbrc.2023.01.050. Epub 2023 Jan 18. Biochem Biophys Res Commun. 2023. PMID: 36689811
-
Caspase-2 activation in the absence of PIDDosome formation.J Cell Biol. 2009 Apr 20;185(2):291-303. doi: 10.1083/jcb.200811105. Epub 2009 Apr 13. J Cell Biol. 2009. PMID: 19364921 Free PMC article.
-
Total recall: the role of PIDDosome components in neurodegeneration.Trends Mol Med. 2023 Dec;29(12):996-1013. doi: 10.1016/j.molmed.2023.08.008. Epub 2023 Sep 14. Trends Mol Med. 2023. PMID: 37716905 Review.
-
The PIDDosome, DNA-damage-induced apoptosis and beyond.Cell Death Differ. 2012 Jan;19(1):13-20. doi: 10.1038/cdd.2011.162. Epub 2011 Nov 18. Cell Death Differ. 2012. PMID: 22095286 Free PMC article. Review.
Cited by
-
Caspase-2 kills cells with extra centrosomes.Sci Adv. 2024 Nov;10(44):eado6607. doi: 10.1126/sciadv.ado6607. Epub 2024 Oct 30. Sci Adv. 2024. PMID: 39475598 Free PMC article.
-
p53 accumulation following cytokinesis failure in the absence of caspase-2.Cell Death Differ. 2018 Nov;25(11):2050-2052. doi: 10.1038/s41418-018-0161-0. Epub 2018 Aug 6. Cell Death Differ. 2018. PMID: 30082771 Free PMC article. No abstract available.
-
Targeting apoptotic caspases in cancer.Biochim Biophys Acta Mol Cell Res. 2020 Jun;1867(6):118688. doi: 10.1016/j.bbamcr.2020.118688. Epub 2020 Feb 19. Biochim Biophys Acta Mol Cell Res. 2020. PMID: 32087180 Free PMC article. Review.
-
Dimer-specific immunoprecipitation of active caspase-2 identifies TRAF proteins as novel activators.EMBO J. 2018 Jul 13;37(14):e97072. doi: 10.15252/embj.201797072. Epub 2018 Jun 6. EMBO J. 2018. PMID: 29875129 Free PMC article.
-
Stepwise phosphorylation and SUMOylation of PIDD1 drive PIDDosome assembly in response to DNA repair failure.Nat Commun. 2024 Oct 24;15(1):9195. doi: 10.1038/s41467-024-53412-0. Nat Commun. 2024. PMID: 39448602 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

