The Msb3/Gyp3 GAP controls the activity of the Rab GTPases Vps21 and Ypt7 at endosomes and vacuoles
- PMID: 22593206
- PMCID: PMC3386215
- DOI: 10.1091/mbc.E11-12-1030
The Msb3/Gyp3 GAP controls the activity of the Rab GTPases Vps21 and Ypt7 at endosomes and vacuoles
Abstract
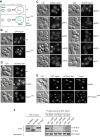
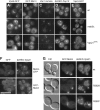
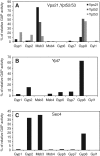
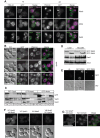
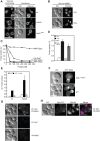
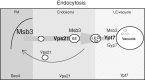
Fusion of organelles in the endomembrane system depends on Rab GTPases that interact with tethering factors before lipid bilayer mixing. In yeast, the Rab5 GTPase Vps21 controls fusion and membrane dynamics between early and late endosomes. Here we identify Msb3/Gyp3 as a specific Vps21 GTPase-activating protein (GAP). Loss of Msb3 results in an accumulation of Vps21 and one of its effectors Vps8, a subunit of the CORVET complex, at the vacuole membrane in vivo. In agreement, Msb3 forms a specific transition complex with Vps21, has the highest activity of all recombinant GAPs for Vps21 in vitro, and is found at vacuoles despite its predominant localization to bud tips and bud necks at the plasma membrane. Surprisingly, Msb3 also inhibits vacuole fusion, which can be rescued by the Ypt7 GDP-GTP exchange factor (GEF), the Mon1-Ccz1 complex. Consistently, msb3 vacuoles fuse more efficiently than wild-type vacuoles in vitro, suggesting that GAP can also act on Ypt7. Our data indicate that GAPs such as Msb3 can act on multiple substrates in vivo at both ends of a trafficking pathway. This ensures specificity of the subsequent GEF-mediated activation of the Rab that initiates the next transport event.
Figures

Similar articles
-
Identification of a Rab GTPase-activating protein cascade that controls recycling of the Rab5 GTPase Vps21 from the vacuole.Mol Biol Cell. 2015 Jul 1;26(13):2535-49. doi: 10.1091/mbc.E15-02-0062. Epub 2015 May 13. Mol Biol Cell. 2015. PMID: 25971802 Free PMC article.
-
The CORVET complex promotes tethering and fusion of Rab5/Vps21-positive membranes.Proc Natl Acad Sci U S A. 2013 Mar 5;110(10):3823-8. doi: 10.1073/pnas.1221785110. Epub 2013 Feb 15. Proc Natl Acad Sci U S A. 2013. PMID: 23417307 Free PMC article.
-
The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7.Curr Biol. 2010 Sep 28;20(18):1654-9. doi: 10.1016/j.cub.2010.08.002. Curr Biol. 2010. PMID: 20797862
-
Membrane dynamics and fusion at late endosomes and vacuoles--Rab regulation, multisubunit tethering complexes and SNAREs.Eur J Cell Biol. 2011 Sep;90(9):779-85. doi: 10.1016/j.ejcb.2011.04.007. Epub 2011 Jun 16. Eur J Cell Biol. 2011. PMID: 21683469 Review.
-
Who's in control? Principles of Rab GTPase activation in endolysosomal membrane trafficking and beyond.J Cell Biol. 2021 Sep 6;220(9):e202105120. doi: 10.1083/jcb.202105120. Epub 2021 Aug 12. J Cell Biol. 2021. PMID: 34383013 Free PMC article. Review.
Cited by
-
Transport mechanisms between the endocytic, recycling, and biosynthetic pathways via endosomes and the trans-Golgi network.Front Cell Dev Biol. 2024 Sep 3;12:1464337. doi: 10.3389/fcell.2024.1464337. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39291266 Free PMC article. Review.
-
The BLOC-1 complex promotes endosomal maturation by recruiting the Rab5 GTPase-activating protein Msb3.J Cell Biol. 2013 Apr 1;201(1):97-111. doi: 10.1083/jcb.201210038. J Cell Biol. 2013. PMID: 23547030 Free PMC article.
-
The yeast endocytic early/sorting compartment exists as an independent sub-compartment within the trans-Golgi network.Elife. 2023 Jul 21;12:e84850. doi: 10.7554/eLife.84850. Elife. 2023. PMID: 37477116 Free PMC article.
-
Rab GAP cascade regulates dynamics of Ypt6 in the Golgi traffic.Proc Natl Acad Sci U S A. 2013 Nov 19;110(47):18976-81. doi: 10.1073/pnas.1308627110. Epub 2013 Nov 5. Proc Natl Acad Sci U S A. 2013. PMID: 24194547 Free PMC article.
-
Rab5-mediated endosome formation is regulated at the trans-Golgi network.Commun Biol. 2019 Nov 15;2:419. doi: 10.1038/s42003-019-0670-5. eCollection 2019. Commun Biol. 2019. PMID: 31754649 Free PMC article.
References
-
- Albert S, Gallwitz D. Two new members of a family of Ypt/Rab GTPase activating proteins. Promiscuity of substrate recognition. J Biol Chem. 1999;274:33186–33189. - PubMed
-
- Albert S, Gallwitz D. Msb4p, a protein involved in Cdc42p-dependent organization of the actin cytoskeleton, is a Ypt/Rab-specific GAP. Biol Chem. 2000;381:453–456. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

