Human transposon tectonics
- PMID: 22579280
- PMCID: PMC3370394
- DOI: 10.1016/j.cell.2012.04.019
Human transposon tectonics
Abstract
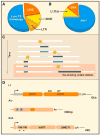
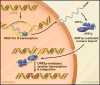
Mobile DNAs have had a central role in shaping our genome. More than half of our DNA is comprised of interspersed repeats resulting from replicative copy and paste events of retrotransposons. Although most are fixed, incapable of templating new copies, there are important exceptions to retrotransposon quiescence. De novo insertions cause genetic diseases and cancers, though reliably detecting these occurrences has been difficult. New technologies aimed at uncovering polymorphic insertions reveal that mobile DNAs provide a substantial and dynamic source of structural variation. Key questions going forward include how and how much new transposition events affect human health and disease.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Natural mutagenesis of human genomes by endogenous retrotransposons.Cell. 2010 Jun 25;141(7):1253-61. doi: 10.1016/j.cell.2010.05.020. Cell. 2010. PMID: 20603005 Free PMC article.
-
A Model-Driven Quantitative Analysis of Retrotransposon Distributions in the Human Genome.Genome Biol Evol. 2020 Nov 3;12(11):2045-2059. doi: 10.1093/gbe/evaa201. Genome Biol Evol. 2020. PMID: 32986810 Free PMC article.
-
Active human retrotransposons: variation and disease.Curr Opin Genet Dev. 2012 Jun;22(3):191-203. doi: 10.1016/j.gde.2012.02.006. Epub 2012 Mar 8. Curr Opin Genet Dev. 2012. PMID: 22406018 Free PMC article. Review.
-
The evolution of mobile DNAs: when will transposons create phylogenies that look as if there is a master gene?Genetics. 2006 Jun;173(2):1115-23. doi: 10.1534/genetics.104.027219. Genetics. 2006. PMID: 16790583 Free PMC article.
-
Retrotransposable elements and human disease.Genome Dyn. 2006;1:104-115. doi: 10.1159/000092503. Genome Dyn. 2006. PMID: 18724056 Review.
Cited by
-
Links between Human LINE-1 Retrotransposons and Hepatitis Virus-Related Hepatocellular Carcinoma.Front Chem. 2016 May 11;4:21. doi: 10.3389/fchem.2016.00021. eCollection 2016. Front Chem. 2016. PMID: 27242996 Free PMC article. Review.
-
Isolation and expression of the human gametocyte-specific factor 1 gene (GTSF1) in fetal ovary, oocytes, and preimplantation embryos.J Assist Reprod Genet. 2017 Jan;34(1):23-31. doi: 10.1007/s10815-016-0795-0. Epub 2016 Sep 19. J Assist Reprod Genet. 2017. PMID: 27646122 Free PMC article.
-
Passport control for foreign integrated DNAs: An unexpected checkpoint by class II HDAC4 revealed by amino acid starvation.Mob Genet Elements. 2012 Sep 1;2(5):233-238. doi: 10.4161/mge.22610. Mob Genet Elements. 2012. PMID: 23550098 Free PMC article.
-
Quantification of LINE-1 RNA Expression from Bulk RNA-seq Using L1EM.Methods Mol Biol. 2023;2607:115-126. doi: 10.1007/978-1-0716-2883-6_7. Methods Mol Biol. 2023. PMID: 36449161
-
RNA sensor MDA5 suppresses LINE-1 retrotransposition by regulating the promoter activity of LINE-1 5'-UTR.Mob DNA. 2022 Apr 12;13(1):10. doi: 10.1186/s13100-022-00268-0. Mob DNA. 2022. PMID: 35414110 Free PMC article.
References
-
- Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. - PubMed
-
- Alves G, Tatro A, Fanning T. Differential methylation of human LINE-1 retrotransposons in malignant cells. Gene. 1996;176:39–44. - PubMed
-
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

