Interaction of the mediator head module with RNA polymerase II
- PMID: 22579255
- PMCID: PMC3383055
- DOI: 10.1016/j.str.2012.02.023
Interaction of the mediator head module with RNA polymerase II
Abstract
Mediator, a large (21 polypeptides, MW ∼1 MDa) complex conserved throughout eukaryotes, plays an essential role in control of gene expression by conveying regulatory signals that influence the activity of the preinitiation complex. However, the precise mode of interaction between Mediator and RNA polymerase II (RNAPII), and the mechanism of regulation by Mediator remain elusive. We used cryo-electron microscopy and reconstituted in vitro transcription assays to characterize a transcriptionally-active complex including the Mediator Head module and components of a minimum preinitiation complex (RNAPII, TFIIF, TFIIB, TBP, and promoter DNA). Our results reveal how the Head interacts with RNAPII, affecting its conformation and function.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures


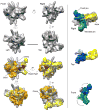
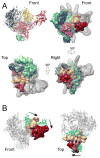


Comment in
-
Mutual remodeling and conformation grid: a mediator code?Structure. 2012 May 9;20(5):755-7. doi: 10.1016/j.str.2012.04.008. Structure. 2012. PMID: 22579244 Free PMC article.
Similar articles
-
Molecular architecture of the human Mediator-RNA polymerase II-TFIIF assembly.PLoS Biol. 2011 Mar;9(3):e1000603. doi: 10.1371/journal.pbio.1000603. Epub 2011 Mar 29. PLoS Biol. 2011. PMID: 21468301 Free PMC article.
-
Mediator head module structure and functional interactions.Nat Struct Mol Biol. 2010 Mar;17(3):273-9. doi: 10.1038/nsmb.1757. Epub 2010 Feb 14. Nat Struct Mol Biol. 2010. PMID: 20154708 Free PMC article.
-
Structure of the human Mediator-RNA polymerase II pre-initiation complex.Nature. 2021 Jun;594(7861):129-133. doi: 10.1038/s41586-021-03555-7. Epub 2021 Apr 26. Nature. 2021. PMID: 33902108
-
More pieces to the puzzle: recent structural insights into class II transcription initiation.Curr Opin Struct Biol. 2014 Feb;24:91-7. doi: 10.1016/j.sbi.2013.12.005. Epub 2014 Jan 16. Curr Opin Struct Biol. 2014. PMID: 24440461 Review.
-
Interactions between subunits of the Mediator complex with gene-specific transcription factors.Semin Cell Dev Biol. 2011 Sep;22(7):759-68. doi: 10.1016/j.semcdb.2011.07.022. Epub 2011 Aug 4. Semin Cell Dev Biol. 2011. PMID: 21839847 Review.
Cited by
-
The Mediator complex and transcription regulation.Crit Rev Biochem Mol Biol. 2013 Nov-Dec;48(6):575-608. doi: 10.3109/10409238.2013.840259. Epub 2013 Oct 3. Crit Rev Biochem Mol Biol. 2013. PMID: 24088064 Free PMC article. Review.
-
Architectural Mediator subunits are differentially essential for global transcription in Saccharomyces cerevisiae.Genetics. 2021 Mar 31;217(3):iyaa042. doi: 10.1093/genetics/iyaa042. Genetics. 2021. PMID: 33789343 Free PMC article.
-
Core promoter-selective coregulators of transcription by RNA polymerase II.Transcription. 2012 Nov-Dec;3(6):295-9. doi: 10.4161/trns.21846. Epub 2012 Nov 1. Transcription. 2012. PMID: 23117823 Free PMC article.
-
Redefining the modular organization of the core Mediator complex.Cell Res. 2014 Jul;24(7):796-808. doi: 10.1038/cr.2014.64. Epub 2014 May 9. Cell Res. 2014. PMID: 24810298 Free PMC article.
-
Mediator is an intrinsic component of the basal RNA polymerase II machinery in vivo.Nucleic Acids Res. 2013 Nov;41(21):9651-62. doi: 10.1093/nar/gkt701. Epub 2013 Aug 20. Nucleic Acids Res. 2013. PMID: 23963697 Free PMC article.
References
-
- Asturias FJ, Jiang YW, Myers LC, Gustafsson CM, Kornberg RD. Conserved structures of mediator and RNA polymerase II holoenzyme. Science. 1999;283:985–987. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

