Profiling Eph receptor expression in cells and tissues: a targeted mass spectrometry approach
- PMID: 22568954
- PMCID: PMC3499309
- DOI: 10.4161/cam.19620
Profiling Eph receptor expression in cells and tissues: a targeted mass spectrometry approach
Abstract
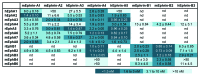
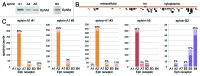
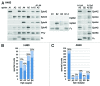
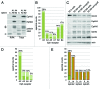
The Eph receptor tyrosine kinase family includes many members, which are often expressed together in various combinations and can promiscuously interact with multiple ephrin ligands, generating intricate networks of intracellular signals that control physiological and pathological processes. Knowing the entire repertoire of Eph receptors and ephrins expressed in a biological sample is important when studying their biological roles. Moreover, given the correlation between Eph receptor/ephrin expression and cancer pathogenesis, their expression patterns could serve important diagnostic and prognostic purposes. However, profiling Eph receptor and ephrin expression has been challenging. Here we describe a novel and straightforward approach to catalog the Eph receptors present in cultured cells and tissues. By measuring the binding of ephrin Fc fusion proteins to Eph receptors in ELISA and pull-down assays, we determined that a mixture of four ephrins is suitable for isolating both EphA and EphB receptors in a single pull-down. We then used mass spectrometry to identify the Eph receptors present in the pull-downs and estimate their relative levels. This approach was validated in cultured human cancer cell lines, human tumor xenograft tissue grown in mice, and mouse brain tissue. The new mass spectrometry approach we have developed represents a useful tool for the identification of the spectrum of Eph receptors present in a biological sample and could also be extended to profiling ephrin expression.
Figures
Similar articles
-
The Functions of EphA1 Receptor Tyrosine Kinase in Several Tumors.Curr Med Chem. 2023;30(20):2340-2353. doi: 10.2174/0929867329666220820125638. Curr Med Chem. 2023. PMID: 35996244 Review.
-
Eph receptor signaling and ephrins.Cold Spring Harb Perspect Biol. 2013 Sep 1;5(9):a009159. doi: 10.1101/cshperspect.a009159. Cold Spring Harb Perspect Biol. 2013. PMID: 24003208 Free PMC article. Review.
-
Expression profile of Eph receptors and ephrin ligands in human skin and downregulation of EphA1 in nonmelanoma skin cancer.Mod Pathol. 2006 Oct;19(10):1369-77. doi: 10.1038/modpathol.3800660. Epub 2006 Jul 7. Mod Pathol. 2006. PMID: 16862074
-
Eph receptor-ephrin bidirectional signals that target Ras and Rho proteins.Cell Signal. 2004 Jun;16(6):655-66. doi: 10.1016/j.cellsig.2003.10.006. Cell Signal. 2004. PMID: 15093606 Review.
-
Drosophila Eph receptor guides specific axon branches of mushroom body neurons.Development. 2006 May;133(9):1845-54. doi: 10.1242/dev.02353. Development. 2006. PMID: 16613832 Free PMC article.
Cited by
-
Eph receptor signalling: from catalytic to non-catalytic functions.Oncogene. 2019 Sep;38(39):6567-6584. doi: 10.1038/s41388-019-0931-2. Epub 2019 Aug 12. Oncogene. 2019. PMID: 31406248 Review.
-
Emerging Roles for Eph Receptors and Ephrin Ligands in Immunity.Front Immunol. 2019 Jul 4;10:1473. doi: 10.3389/fimmu.2019.01473. eCollection 2019. Front Immunol. 2019. PMID: 31333644 Free PMC article. Review.
-
Transmembrane helix interactions regulate oligomerization of the receptor tyrosine kinase EphA2.J Biol Chem. 2024 Jul;300(7):107441. doi: 10.1016/j.jbc.2024.107441. Epub 2024 Jun 3. J Biol Chem. 2024. PMID: 38838777 Free PMC article.
-
Protein kinase A can block EphA2 receptor-mediated cell repulsion by increasing EphA2 S897 phosphorylation.Mol Biol Cell. 2016 Sep 1;27(17):2757-70. doi: 10.1091/mbc.E16-01-0048. Epub 2016 Jul 6. Mol Biol Cell. 2016. PMID: 27385333 Free PMC article.
-
Actomyosin regulation by Eph receptor signaling couples boundary cell formation to border sharpness.Elife. 2019 Sep 10;8:e49696. doi: 10.7554/eLife.49696. Elife. 2019. PMID: 31502954 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
