Spatial Organization and Dynamics of Transcription Elongation and Pre-mRNA Processing in Live Cells
- PMID: 22567362
- PMCID: PMC3335512
- DOI: 10.4061/2011/626081
Spatial Organization and Dynamics of Transcription Elongation and Pre-mRNA Processing in Live Cells
Abstract
During the last 30 years, systematic biochemical and functional studies have significantly expanded our knowledge of the transcriptional molecular components and the pre-mRNA processing machinery of the cell. However, our current understanding of how these functions take place spatiotemporally within the highly compartmentalized eukaryotic nucleus remains limited. Moreover, it is increasingly clear that "the whole is more than the sum of its parts" and that an understanding of the dynamic coregulation of genes is essential for fully characterizing complex biological phenomena and underlying diseases. Recent technological advances in light microscopy in addition to novel cell and molecular biology approaches have led to the development of new tools, which are being used to address these questions and may contribute to achieving an integrated and global understanding of how the genome works at a cellular level. Here, we review major hallmarks and novel insights in RNA polymerase II activity and pre-mRNA processing in the context of nuclear organization, as well as new concepts and challenges arising from our ability to gather extensive dynamic information at the single-cell resolution.
Figures
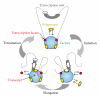
Similar articles
-
Functional coupling of transcription and splicing.Gene. 2012 Jun 15;501(2):104-17. doi: 10.1016/j.gene.2012.04.006. Epub 2012 Apr 17. Gene. 2012. PMID: 22537677 Review.
-
Targeting proteins to RNA transcription and processing sites within the nucleus.Int J Biochem Cell Biol. 2017 Oct;91(Pt B):194-202. doi: 10.1016/j.biocel.2017.06.001. Epub 2017 Jun 6. Int J Biochem Cell Biol. 2017. PMID: 28600144 Review.
-
Primary transcripts: From the discovery of RNA processing to current concepts of gene expression - Review.Exp Cell Res. 2018 Dec 15;373(1-2):1-33. doi: 10.1016/j.yexcr.2018.09.011. Epub 2018 Sep 26. Exp Cell Res. 2018. PMID: 30266658 Review.
-
News from Mars: Two-Tier Paradox, Intracellular PCR, Chimeric Junction Shift, Dark Matter mRNA and Other Remarkable Features of Mammalian RNA-Dependent mRNA Amplification. Implications for Alzheimer's Disease, RNA-Based Vaccines and mRNA Therapeutics.Ann Integr Mol Med. 2021;2:131-173. doi: 10.33597/aimm.02-1009. Ann Integr Mol Med. 2021. PMID: 33942036 Free PMC article.
-
Dynamics of the mammalian nucleus: can microscopic movements help us to understand our genes?Philos Trans A Math Phys Eng Sci. 2004 Dec 15;362(1825):2775-93. doi: 10.1098/rsta.2004.1463. Philos Trans A Math Phys Eng Sci. 2004. PMID: 15539370 Review.
References
-
- Carmo-Fonseca M, Carvalho C. Nuclear organization and splicing control. Advances in Experimental Medicine and Biology. 2007;623:1–13. - PubMed
-
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nature Reviews Genetics. 2001;2(4):292–301. - PubMed
-
- Parada L, Misteli T. Chromosome positioning in the interphase nucleus. Trends in Cell Biology. 2002;12(9):425–432. - PubMed

