Prion formation by a yeast GLFG nucleoporin
- PMID: 22561191
- PMCID: PMC3609069
- DOI: 10.4161/pri.20199
Prion formation by a yeast GLFG nucleoporin
Abstract
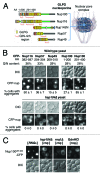
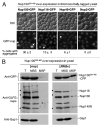
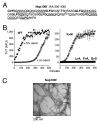
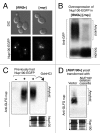
The self-assembly of proteins into higher order structures is both central to normal biology and a dominant force in disease. Certain glutamine/asparagine (Q/N)-rich proteins in the budding yeast Saccharomyces cerevisiae assemble into self-replicating amyloid-like protein polymers, or prions, that act as genetic elements in an entirely protein-based system of inheritance. The nuclear pore complex (NPC) contains multiple Q/N-rich proteins whose self-assembly has also been proposed to underlie structural and functional properties of the NPC. Here we show that an essential sequence feature of these proteins--repeating GLFG motifs--strongly promotes their self-assembly into amyloids with characteristics of prions. Furthermore, we demonstrate that Nup100 can form bona fide prions, thus establishing a previously undiscovered ability of yeast GLFG nucleoporins to adopt this conformational state in vivo.
Figures

Similar articles
-
Entry into the nuclear pore complex is controlled by a cytoplasmic exclusion zone containing dynamic GLFG-repeat nucleoporin domains.J Cell Sci. 2014 Jan 1;127(Pt 1):124-36. doi: 10.1242/jcs.133272. Epub 2013 Oct 21. J Cell Sci. 2014. PMID: 24144701
-
Opposing effects of glutamine and asparagine govern prion formation by intrinsically disordered proteins.Mol Cell. 2011 Jul 8;43(1):72-84. doi: 10.1016/j.molcel.2011.05.013. Mol Cell. 2011. PMID: 21726811 Free PMC article.
-
Natively Unfolded FG Repeats Stabilize the Structure of the Nuclear Pore Complex.Cell. 2017 Nov 2;171(4):904-917.e19. doi: 10.1016/j.cell.2017.09.033. Epub 2017 Oct 12. Cell. 2017. PMID: 29033133 Free PMC article.
-
Life cycle of yeast prions: propagation mediated by amyloid fibrils.Protein Pept Lett. 2009;16(3):271-6. doi: 10.2174/092986609787601796. Protein Pept Lett. 2009. PMID: 19275740 Review.
-
Prion amyloid structure explains templating: how proteins can be genes.FEMS Yeast Res. 2010 Dec;10(8):980-91. doi: 10.1111/j.1567-1364.2010.00666.x. FEMS Yeast Res. 2010. PMID: 20726897 Free PMC article. Review.
Cited by
-
Conservation of Prion-Like Composition and Sequence in Prion-Formers and Prion-Like Proteins of Saccharomyces cerevisiae.Front Mol Biosci. 2019 Jul 11;6:54. doi: 10.3389/fmolb.2019.00054. eCollection 2019. Front Mol Biosci. 2019. PMID: 31355208 Free PMC article.
-
Disordered proteinaceous machines.Chem Rev. 2014 Jul 9;114(13):6806-43. doi: 10.1021/cr4007329. Epub 2014 Apr 4. Chem Rev. 2014. PMID: 24702702 Free PMC article. Review. No abstract available.
-
Defining the limits: Protein aggregation and toxicity in vivo.Crit Rev Biochem Mol Biol. 2014 Jul-Aug;49(4):294-303. doi: 10.3109/10409238.2014.914151. Epub 2014 Apr 28. Crit Rev Biochem Mol Biol. 2014. PMID: 24766537 Free PMC article. Review.
-
Systematic analysis of barrier-forming FG hydrogels from Xenopus nuclear pore complexes.EMBO J. 2013 Jan 23;32(2):204-18. doi: 10.1038/emboj.2012.302. Epub 2012 Nov 30. EMBO J. 2013. PMID: 23202855 Free PMC article.
-
Saccharomyces cerevisiae in neuroscience: how unicellular organism helps to better understand prion protein?Neural Regen Res. 2021 Mar;16(3):489-495. doi: 10.4103/1673-5374.293137. Neural Regen Res. 2021. PMID: 32985470 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
