Regulation of histone gene expression in budding yeast
- PMID: 22555441
- PMCID: PMC3338271
- DOI: 10.1534/genetics.112.140145
Regulation of histone gene expression in budding yeast
Abstract
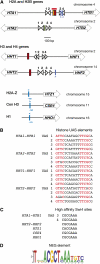
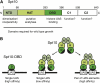
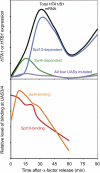
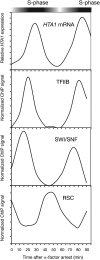
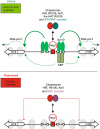
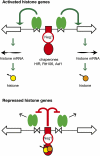
We discuss the regulation of the histone genes of the budding yeast Saccharomyces cerevisiae. These include genes encoding the major core histones (H3, H4, H2A, and H2B), histone H1 (HHO1), H2AZ (HTZ1), and centromeric H3 (CSE4). Histone production is regulated during the cell cycle because the cell must replicate both its DNA during S phase and its chromatin. Consequently, the histone genes are activated in late G1 to provide sufficient core histones to assemble the replicated genome into chromatin. The major core histone genes are subject to both positive and negative regulation. The primary control system is positive, mediated by the histone gene-specific transcription activator, Spt10, through the histone upstream activating sequences (UAS) elements, with help from the major G1/S-phase activators, SBF (Swi4 cell cycle box binding factor) and perhaps MBF (MluI cell cycle box binding factor). Spt10 binds specifically to the histone UAS elements and contains a putative histone acetyltransferase domain. The negative system involves negative regulatory elements in the histone promoters, the RSC chromatin-remodeling complex, various histone chaperones [the histone regulatory (HIR) complex, Asf1, and Rtt106], and putative sequence-specific factors. The SWI/SNF chromatin-remodeling complex links the positive and negative systems. We propose that the negative system is a damping system that modulates the amount of transcription activated by Spt10 and SBF. We hypothesize that the negative system mediates negative feedback on the histone genes by histone proteins through the level of saturation of histone chaperones with histone. Thus, the negative system could communicate the degree of nucleosome assembly during DNA replication and the need to shut down the activating system under replication-stress conditions. We also discuss post-transcriptional regulation and dosage compensation of the histone genes.
Figures
Similar articles
-
Spt10 and Swi4 control the timing of histone H2A/H2B gene activation in budding yeast.Mol Cell Biol. 2011 Feb;31(3):557-72. doi: 10.1128/MCB.00909-10. Epub 2010 Nov 29. Mol Cell Biol. 2011. PMID: 21115727 Free PMC article.
-
The Saccharomyces cerevisiae histone chaperone Rtt106 mediates the cell cycle recruitment of SWI/SNF and RSC to the HIR-dependent histone genes.PLoS One. 2011;6(6):e21113. doi: 10.1371/journal.pone.0021113. Epub 2011 Jun 15. PLoS One. 2011. PMID: 21698254 Free PMC article.
-
The mitotic Clb cyclins are required to alleviate HIR-mediated repression of the yeast histone genes at the G1/S transition.Biochim Biophys Acta. 2012 Jan;1819(1):16-27. doi: 10.1016/j.bbagrm.2011.09.003. Epub 2011 Sep 28. Biochim Biophys Acta. 2012. PMID: 21978826 Free PMC article.
-
Regulation of histone gene transcription in yeast.Cell Mol Life Sci. 2014 Feb;71(4):599-613. doi: 10.1007/s00018-013-1443-9. Epub 2013 Aug 23. Cell Mol Life Sci. 2014. PMID: 23974242 Free PMC article. Review.
-
Histone post-translational modifications regulate transcription and silent chromatin in Saccharomyces cerevisiae.Ernst Schering Res Found Workshop. 2006;(57):127-53. doi: 10.1007/3-540-37633-x_8. Ernst Schering Res Found Workshop. 2006. PMID: 16568953 Review.
Cited by
-
The fate of the messenger is pre-determined: a new model for regulation of gene expression.Biochim Biophys Acta. 2013 Jun-Jul;1829(6-7):643-53. doi: 10.1016/j.bbagrm.2013.01.004. Epub 2013 Jan 19. Biochim Biophys Acta. 2013. PMID: 23337853 Free PMC article. Review.
-
Resetting the Yeast Epigenome with Human Nucleosomes.Cell. 2017 Dec 14;171(7):1508-1519.e13. doi: 10.1016/j.cell.2017.10.043. Epub 2017 Nov 30. Cell. 2017. PMID: 29198523 Free PMC article.
-
A novel approach for studying histone H1 function in vivo.Genetics. 2015 May;200(1):29-33. doi: 10.1534/genetics.114.170514. Epub 2015 Mar 23. Genetics. 2015. PMID: 25805849 Free PMC article.
-
High levels of histones promote whole-genome-duplications and trigger a Swe1WEE1-dependent phosphorylation of Cdc28CDK1.Elife. 2018 Mar 27;7:e35337. doi: 10.7554/eLife.35337. Elife. 2018. PMID: 29580382 Free PMC article.
-
The functional study of human proteins using humanized yeast.J Microbiol. 2020 May;58(5):343-349. doi: 10.1007/s12275-020-0136-y. Epub 2020 Apr 27. J Microbiol. 2020. PMID: 32342338 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

