Wnt5a participates in hepatic stellate cell activation observed by gene expression profile and functional assays
- PMID: 22553398
- PMCID: PMC3332287
- DOI: 10.3748/wjg.v18.i15.1745
Wnt5a participates in hepatic stellate cell activation observed by gene expression profile and functional assays
Abstract
Aim: To identify differentially expressed genes in quiescent and activated hepatic stellate cells (HSCs) and explore their functions.
Methods: HSCs were isolated from the normal Sprague Dawley rats by in suit perfusion of collagenase and pronase and density Nycodenz gradient centrifugation. Total RNA and mRNA of quiescent HSCs, and culture-activated HSCs were extracted, quantified and reversely transcripted into cDNA. The global gene expression profile was analyzed by microarray with Affymetrix rat genechip. Differentially expressed genes were annotated with Gene Ontology (GO) and analyzed with Kyoto encyclopedia of genes and genomes (KEGG) pathway using the Database for Annotation, Visualization and Integrated Discovery. Microarray data were validated by quantitative real-time polymerase chain reaction (qRT-PCR). The function of Wnt5a on human HSCs line LX-2 was assessed with lentivirus-mediated Wnt5a RNAi. The expression of Wnt5a in fibrotic liver of a carbon tetrachloride (CCl(4))-induced fibrosis rat model was also analyzed with Western blotting.
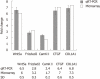
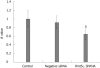
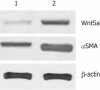
Results: Of the 28 700 genes represented on this chip, 2566 genes displayed at least a 2-fold increase or decrease in expression at a P < 0.01 level with a false discovery rate. Of these, 1396 genes were upregulated, while 1170 genes were downregulated in culture-activated HSCs. These differentially expressed transcripts were grouped into 545 GO based on biological process GO terms. The most enriched GO terms included response to wounding, wound healing, regulation of cell growth, vasculature development and actin cytoskeleton organization. KEGG pathway analysis revealed that Wnt5a signaling pathway participated in the activation of HSCs. Wnt5a was significantly increased in culture-activated HSCs as compared with quiescent HSCs. qRT-PCR validated the microarray data. Lentivirus-mediated suppression of Wnt5a expression in activated LX-2 resulted in significantly impaired proliferation, downregulated expressions of type I collagen and transforming growth factor-β1. Wnt5a was upregulated in the fibrotic liver of a CCl(4)-induced fibrosis rat model.
Conclusion: Wnt5a is involved in the activation of HSCs, and it may serve as a novel therapeutic target in the treatment of liver fibrosis.
Keywords: Bioinformatics analyses; Hepatic stellate cells; Liver fibrosis; Microarray; Wnt5a.
Figures



Similar articles
-
Gene expression profile of quiescent and activated rat hepatic stellate cells implicates Wnt signaling pathway in activation.J Hepatol. 2006 Sep;45(3):401-9. doi: 10.1016/j.jhep.2006.03.016. Epub 2006 May 3. J Hepatol. 2006. PMID: 16780995
-
Downregulation of the Wnt antagonist Dkk2 links the loss of Sept4 and myofibroblastic transformation of hepatic stellate cells.Biochim Biophys Acta. 2011 Nov;1812(11):1403-11. doi: 10.1016/j.bbadis.2011.06.015. Epub 2011 Jul 6. Biochim Biophys Acta. 2011. PMID: 21763422
-
RNA Sequencing and Bioinformatics Analysis Implicate the Regulatory Role of a Long Noncoding RNA-mRNA Network in Hepatic Stellate Cell Activation.Cell Physiol Biochem. 2017;42(5):2030-2042. doi: 10.1159/000479898. Epub 2017 Aug 11. Cell Physiol Biochem. 2017. PMID: 28803234
-
Wnt signaling in liver fibrosis: progress, challenges and potential directions.Biochimie. 2013 Dec;95(12):2326-35. doi: 10.1016/j.biochi.2013.09.003. Epub 2013 Sep 13. Biochimie. 2013. PMID: 24036368 Review.
-
Hepatic stellate cells: partners in crime for liver metastases?Hepatology. 2011 Aug;54(2):707-13. doi: 10.1002/hep.24384. Hepatology. 2011. PMID: 21520207 Free PMC article. Review.
Cited by
-
Differentially expressed mRNAs and lncRNAs shared between activated human hepatic stellate cells and nash fibrosis.Biochem Biophys Rep. 2020 Mar 24;22:100753. doi: 10.1016/j.bbrep.2020.100753. eCollection 2020 Jul. Biochem Biophys Rep. 2020. PMID: 32258441 Free PMC article.
-
Epigenetic Mechanisms in Hepatic Stellate Cell Activation During Liver Fibrosis and Carcinogenesis.Int J Mol Sci. 2019 May 21;20(10):2507. doi: 10.3390/ijms20102507. Int J Mol Sci. 2019. PMID: 31117267 Free PMC article. Review.
-
RNA sequencing of LX-2 cells treated with TGF-β1 identifies genes associated with hepatic stellate cell activation.Mol Biol Rep. 2021 Dec;48(12):7677-7688. doi: 10.1007/s11033-021-06774-3. Epub 2021 Oct 14. Mol Biol Rep. 2021. PMID: 34648138 Free PMC article.
-
β-Catenin Signaling and Roles in Liver Homeostasis, Injury, and Tumorigenesis.Gastroenterology. 2015 Jun;148(7):1294-310. doi: 10.1053/j.gastro.2015.02.056. Epub 2015 Mar 5. Gastroenterology. 2015. PMID: 25747274 Free PMC article. Review.
-
Differential gene expression of Wnt signaling pathway in benign, premalignant, and malignant human breast epithelial cells.Tumour Biol. 2012 Dec;33(6):2317-27. doi: 10.1007/s13277-012-0494-0. Epub 2012 Sep 1. Tumour Biol. 2012. PMID: 22941468
References
-
- Atzori L, Poli G, Perra A. Hepatic stellate cell: a star cell in the liver. Int J Biochem Cell Biol. 2009;41:1639–1642. - PubMed
-
- Li JT, Liao ZX, Ping J, Xu D, Wang H. Molecular mechanism of hepatic stellate cell activation and antifibrotic therapeutic strategies. J Gastroenterol. 2008;43:419–428. - PubMed
-
- De Minicis S, Seki E, Uchinami H, Kluwe J, Zhang Y, Brenner DA, Schwabe RF. Gene expression profiles during hepatic stellate cell activation in culture and in vivo. Gastroenterology. 2007;132:1937–1946. - PubMed
-
- Woo SW, Hwang KI, Chung MW, Jin SK, Bang S, Lee SH, Lee SH, Chung HJ, Sohn DH. Gene expression profiles during the activation of rat hepatic stellate cells evaluated by cDNA microarray. Arch Pharm Res. 2007;30:1410–1418. - PubMed
-
- Sancho-Bru P, Bataller R, Gasull X, Colmenero J, Khurdayan V, Gual A, Nicolás JM, Arroyo V, Ginès P. Genomic and functional characterization of stellate cells isolated from human cirrhotic livers. J Hepatol. 2005;43:272–282. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

