Cell survival, DNA damage, and oncogenic transformation after a transient and reversible apoptotic response
- PMID: 22535522
- PMCID: PMC3374744
- DOI: 10.1091/mbc.E11-11-0926
Cell survival, DNA damage, and oncogenic transformation after a transient and reversible apoptotic response
Abstract
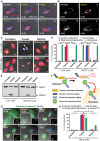
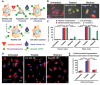
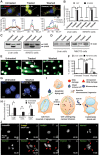
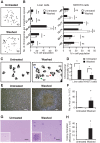
Apoptosis serves as a protective mechanism by eliminating damaged cells through programmed cell death. After apoptotic cells pass critical checkpoints, including mitochondrial fragmentation, executioner caspase activation, and DNA damage, it is assumed that cell death inevitably follows. However, this assumption has not been tested directly. Here we report an unexpected reversal of late-stage apoptosis in primary liver and heart cells, macrophages, NIH 3T3 fibroblasts, cervical cancer HeLa cells, and brain cells. After exposure to an inducer of apoptosis, cells exhibited multiple morphological and biochemical hallmarks of late-stage apoptosis, including mitochondrial fragmentation, caspase-3 activation, and DNA damage. Surprisingly, the vast majority of dying cells arrested the apoptotic process and recovered when the inducer was washed away. Of importance, some cells acquired permanent genetic changes and underwent oncogenic transformation at a higher frequency than controls. Global gene expression analysis identified a molecular signature of the reversal process. We propose that reversal of apoptosis is an unanticipated mechanism to rescue cells from crisis and propose to name this mechanism "anastasis" (Greek for "rising to life"). Whereas carcinogenesis represents a harmful side effect, potential benefits of anastasis could include preservation of cells that are difficult to replace and stress-induced genetic diversity.
Figures
Similar articles
-
Strategies for tracking anastasis, a cell survival phenomenon that reverses apoptosis.J Vis Exp. 2015 Feb 16;(96):51964. doi: 10.3791/51964. J Vis Exp. 2015. PMID: 25742050 Free PMC article.
-
Transcriptomic study of anastasis for reversal of ethanol-induced apoptosis in mouse primary liver cells.Sci Data. 2022 Jul 18;9(1):418. doi: 10.1038/s41597-022-01470-8. Sci Data. 2022. PMID: 35851273 Free PMC article.
-
Molecular profiling of anastatic cancer cells: potential role of the nuclear export pathway.Cell Oncol (Dordr). 2019 Oct;42(5):645-661. doi: 10.1007/s13402-019-00451-1. Epub 2019 May 30. Cell Oncol (Dordr). 2019. PMID: 31147963
-
Integration of EMT and cellular survival instincts in reprogramming of programmed cell death to anastasis.Cancer Metastasis Rev. 2020 Jun;39(2):553-566. doi: 10.1007/s10555-020-09866-x. Cancer Metastasis Rev. 2020. PMID: 32020420 Review.
-
Anastasis: cell recovery mechanisms and potential role in cancer.Cell Commun Signal. 2022 Jun 3;20(1):81. doi: 10.1186/s12964-022-00880-w. Cell Commun Signal. 2022. PMID: 35659306 Free PMC article. Review.
Cited by
-
Therapeutic Synergy in Esophageal Cancer and Mesothelioma Is Predicted by Dynamic BH3 Profiling.Mol Cancer Ther. 2021 Aug;20(8):1469-1480. doi: 10.1158/1535-7163.MCT-20-0887. Epub 2021 Jun 4. Mol Cancer Ther. 2021. PMID: 34088830 Free PMC article.
-
Chromothripsis-Explosion in Genetic Science.Cells. 2021 May 4;10(5):1102. doi: 10.3390/cells10051102. Cells. 2021. PMID: 34064429 Free PMC article. Review.
-
Mitochondria as multifaceted regulators of cell death.Nat Rev Mol Cell Biol. 2020 Feb;21(2):85-100. doi: 10.1038/s41580-019-0173-8. Epub 2019 Oct 21. Nat Rev Mol Cell Biol. 2020. PMID: 31636403 Review.
-
Rapid coupling between gravitational forces and the transcriptome in human myelomonocytic U937 cells.Sci Rep. 2018 Sep 5;8(1):13267. doi: 10.1038/s41598-018-31596-y. Sci Rep. 2018. PMID: 30185876 Free PMC article.
-
Sublethal engagement of apoptotic pathways in residual cancer.Trends Cell Biol. 2024 Mar;34(3):225-238. doi: 10.1016/j.tcb.2023.07.005. Epub 2023 Aug 10. Trends Cell Biol. 2024. PMID: 37573235 Free PMC article. Review.
References
-
- Aitken RJ, Findlay JK, Hutt KJ, Kerr JB. Apoptosis in the germ line. Reproduction. 2011;141:139–150. - PubMed
-
- Arama E, Agapite J, Steller H. Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev Cell. 2003;4:687–697. - PubMed
-
- Bloom AD. Induced chromosomal aberrations: biological and clinical significance. J Pediatr. 1972;81:1–8. - PubMed
-
- Boffetta P, Hashibe M. Alcohol and cancer. Lancet Oncol. 2006;7:149–156. - PubMed
-
- Capy P, Gasperi G, Biemont C, Bazin C. Stress and transposable elements: co-evolution or useful parasites? Heredity. 2000;85:101–106. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

