Tyrosyl-DNA phosphodiesterase 1 initiates repair of apurinic/apyrimidinic sites
- PMID: 22522093
- PMCID: PMC3778944
- DOI: 10.1016/j.biochi.2012.04.004
Tyrosyl-DNA phosphodiesterase 1 initiates repair of apurinic/apyrimidinic sites
Abstract
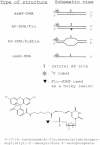
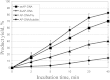
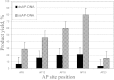
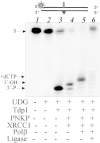
Tyrosyl-DNA phosphodiesterase 1 (Tdp1) catalyzes the hydrolysis of the phosphodiester linkage between the DNA 3' phosphate and a tyrosine residue as well as a variety of other DNA 3' damaged termini. Recently we have shown that Tdp1 can liberate the 3' DNA phosphate termini from apurinic/apyrimidinic (AP) sites. Here, we found that Tdp1 is more active in the cleavage of the AP sites inside bubble-DNA structure in comparison to ssDNA containing AP site. Furthermore, Tdp1 hydrolyzes AP sites opposite to bulky fluorescein adduct faster than AP sites located in dsDNA. Whilst the Tdp1 H493R (SCAN1) and H263A mutants retain the ability to bind an AP site-containing DNA, both mutants do not reveal endonuclease activity, further suggesting the specificity of the AP cleavage activity. We suggest that this Tdp1 activity can contribute to the repair of AP sites particularly in DNA structures containing ssDNA region or AP sites in the context of clustered DNA lesions.
Copyright © 2012 Elsevier Masson SAS. All rights reserved.
Figures
Similar articles
-
Pre-steady state kinetics of DNA binding and abasic site hydrolysis by tyrosyl-DNA phosphodiesterase 1.J Biomol Struct Dyn. 2017 Aug;35(11):2314-2327. doi: 10.1080/07391102.2016.1220331. Epub 2016 Aug 16. J Biomol Struct Dyn. 2017. PMID: 27687298
-
[Tyrosyl-DNA Phosphodiesterase 1 Is a New Player in Repair of Apurinic/Apyrimidinic Sites].Bioorg Khim. 2015 Sep-Oct;41(5):531-8. doi: 10.1134/s106816201505012x. Bioorg Khim. 2015. PMID: 26762090 Review. Russian.
-
The mechanism of human tyrosyl-DNA phosphodiesterase 1 in the cleavage of AP site and its synthetic analogs.DNA Repair (Amst). 2013 Dec;12(12):1037-42. doi: 10.1016/j.dnarep.2013.09.008. Epub 2013 Oct 31. DNA Repair (Amst). 2013. PMID: 24183900
-
Design of a New Fluorescent Oligonucleotide-Based Assay for a Highly Specific Real-Time Detection of Apurinic/Apyrimidinic Site Cleavage by Tyrosyl-DNA Phosphodiesterase 1.Bioconjug Chem. 2015 Oct 21;26(10):2046-53. doi: 10.1021/acs.bioconjchem.5b00451. Epub 2015 Sep 10. Bioconjug Chem. 2015. PMID: 26335988
-
Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3' ends justify the means.Mutat Res. 2000 Aug 30;460(3-4):211-29. doi: 10.1016/s0921-8777(00)00028-8. Mutat Res. 2000. PMID: 10946230 Review.
Cited by
-
Abacavir, an anti-HIV-1 drug, targets TDP1-deficient adult T cell leukemia.Sci Adv. 2015 Apr 24;1(3):e1400203. doi: 10.1126/sciadv.1400203. eCollection 2015 Apr. Sci Adv. 2015. PMID: 26601161 Free PMC article.
-
Dysregulated human Tyrosyl-DNA phosphodiesterase I acts as cellular toxin.Oncotarget. 2016 Dec 27;7(52):86660-86674. doi: 10.18632/oncotarget.13528. Oncotarget. 2016. PMID: 27893431 Free PMC article.
-
A mechanism for oxidative damage repair at gene regulatory elements.Nature. 2022 Sep;609(7929):1038-1047. doi: 10.1038/s41586-022-05217-8. Epub 2022 Sep 28. Nature. 2022. PMID: 36171374
-
Mitochondrial protein-linked DNA breaks perturb mitochondrial gene transcription and trigger free radical-induced DNA damage.Sci Adv. 2017 Apr 28;3(4):e1602506. doi: 10.1126/sciadv.1602506. eCollection 2017 Apr. Sci Adv. 2017. PMID: 28508041 Free PMC article.
-
Resolution of complex ends by Nonhomologous end joining - better to be lucky than good?Genome Integr. 2012 Dec 31;3(1):10. doi: 10.1186/2041-9414-3-10. Genome Integr. 2012. PMID: 23276302 Free PMC article.
References
-
- Pouliot J.J., Robertson C.A., Nash H.A. Pathways for repair of topoisomerase I covalent complexes in Saccharomyces cerevisiae. Genes Cells. 2001;6:677–687. - PubMed
-
- Raymond A.C., Staker B.L., Burgin A.B., Jr. Substrate specificity of tyrosyl-DNA phosphodiesterase I (Tdp1) J. Biol. Chem. 2005;280:22029–22035. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

