Aggrephagy: selective disposal of protein aggregates by macroautophagy
- PMID: 22518139
- PMCID: PMC3320095
- DOI: 10.1155/2012/736905
Aggrephagy: selective disposal of protein aggregates by macroautophagy
Abstract
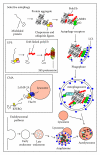
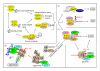
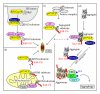
Protein aggregation is a continuous process in our cells. Some proteins aggregate in a regulated manner required for different vital functional processes in the cells whereas other protein aggregates result from misfolding caused by various stressors. The decision to form an aggregate is largely made by chaperones and chaperone-assisted proteins. Proteins that are damaged beyond repair are degraded either by the proteasome or by the lysosome via autophagy. The aggregates can be degraded by the proteasome and by chaperone-mediated autophagy only after dissolution into soluble single peptide species. Hence, protein aggregates as such are degraded by macroautophagy. The selective degradation of protein aggregates by macroautophagy is called aggrephagy. Here we review the processes of aggregate formation, recognition, transport, and sequestration into autophagosomes by autophagy receptors and the role of aggrephagy in different protein aggregation diseases.
Figures
Similar articles
-
Aggrephagy Deficiency in the Placenta: A New Pathogenesis of Preeclampsia.Int J Mol Sci. 2021 Feb 28;22(5):2432. doi: 10.3390/ijms22052432. Int J Mol Sci. 2021. PMID: 33670947 Free PMC article. Review.
-
Aggrephagy at a glance.J Cell Sci. 2023 May 15;136(10):jcs260888. doi: 10.1242/jcs.260888. Epub 2023 May 31. J Cell Sci. 2023. PMID: 37254869 Review.
-
Direct observation of aggregate-triggered selective autophagy in human cells.J Cell Sci. 2021 Oct 1;134(19):jcs258824. doi: 10.1242/jcs.258824. Epub 2021 Oct 6. J Cell Sci. 2021. PMID: 34447998 Free PMC article.
-
Phase Separation in Regulation of Aggrephagy.J Mol Biol. 2020 Jan 3;432(1):160-169. doi: 10.1016/j.jmb.2019.06.026. Epub 2019 Jun 28. J Mol Biol. 2020. PMID: 31260696 Review.
-
A switch of chaperonin function regulates the clearance of solid protein aggregates.Autophagy. 2022 Nov;18(11):2746-2748. doi: 10.1080/15548627.2022.2052581. Epub 2022 Apr 5. Autophagy. 2022. PMID: 35380909 Free PMC article.
Cited by
-
Selective autophagy as a therapeutic target for neurological diseases.Cell Mol Life Sci. 2021 Feb;78(4):1369-1392. doi: 10.1007/s00018-020-03667-9. Epub 2020 Oct 16. Cell Mol Life Sci. 2021. PMID: 33067655 Free PMC article. Review.
-
Autophagy: a crucial moderator of redox balance, inflammation, and apoptosis in lung disease.Antioxid Redox Signal. 2014 Jan 20;20(3):474-94. doi: 10.1089/ars.2013.5373. Epub 2013 Sep 26. Antioxid Redox Signal. 2014. PMID: 23879400 Free PMC article. Review.
-
TRIM16 controls assembly and degradation of protein aggregates by modulating the p62-NRF2 axis and autophagy.EMBO J. 2018 Sep 14;37(18):e98358. doi: 10.15252/embj.201798358. Epub 2018 Aug 24. EMBO J. 2018. PMID: 30143514 Free PMC article.
-
Komagataella phaffii Cue5 Piggybacks on Lipid Droplets for Its Vacuolar Degradation during Stationary Phase Lipophagy.Cells. 2022 Jan 10;11(2):215. doi: 10.3390/cells11020215. Cells. 2022. PMID: 35053331 Free PMC article.
-
Autophagy is induced and modulated by cholesterol depletion through transcription of autophagy-related genes and attenuation of flux.Cell Death Discov. 2021 Oct 29;7(1):320. doi: 10.1038/s41420-021-00718-3. Cell Death Discov. 2021. PMID: 34716312 Free PMC article.
References
-
- Dobson CM. Protein folding and misfolding. Nature. 2003;426(6968):884–890. - PubMed
-
- Komander D. The emerging complexity of protein ubiquitination. Biochemical Society Transactions. 2009;37(5):937–953. - PubMed
-
- Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. Journal of Biological Chemistry. 1983;258(13):8206–8214. - PubMed
LinkOut - more resources
Full Text Sources

