Mitogen-activated protein kinase signaling in plant-interacting fungi: distinct messages from conserved messengers
- PMID: 22517321
- PMCID: PMC3398478
- DOI: 10.1105/tpc.112.096156
Mitogen-activated protein kinase signaling in plant-interacting fungi: distinct messages from conserved messengers
Abstract
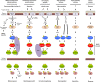
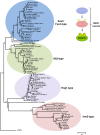
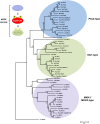
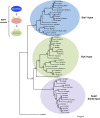
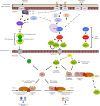
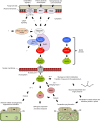
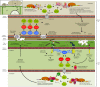
Mitogen-activated protein kinases (MAPKs) are evolutionarily conserved proteins that function as key signal transduction components in fungi, plants, and mammals. During interaction between phytopathogenic fungi and plants, fungal MAPKs help to promote mechanical and/or enzymatic penetration of host tissues, while plant MAPKs are required for activation of plant immunity. However, new insights suggest that MAPK cascades in both organisms do not operate independently but that they mutually contribute to a highly interconnected molecular dialogue between the plant and the fungus. As a result, some pathogenesis-related processes controlled by fungal MAPKs lead to the activation of plant signaling, including the recruitment of plant MAPK cascades. Conversely, plant MAPKs promote defense mechanisms that threaten the survival of fungal cells, leading to a stress response mediated in part by fungal MAPK cascades. In this review, we make use of the genomic data available following completion of whole-genome sequencing projects to analyze the structure of MAPK protein families in 24 fungal taxa, including both plant pathogens and mycorrhizal symbionts. Based on conserved patterns of sequence diversification, we also propose the adoption of a unified fungal MAPK nomenclature derived from that established for the model species Saccharomyces cerevisiae. Finally, we summarize current knowledge of the functions of MAPK cascades in phytopathogenic fungi and highlight the central role played by MAPK signaling during the molecular dialogue between plants and invading fungal pathogens.
Figures
Similar articles
-
MAPK cascades in plant disease resistance signaling.Annu Rev Phytopathol. 2013;51:245-66. doi: 10.1146/annurev-phyto-082712-102314. Epub 2013 May 6. Annu Rev Phytopathol. 2013. PMID: 23663002 Review.
-
Functional analysis of the MAPK pathways in fungi.Rev Iberoam Micol. 2017 Oct-Dec;34(4):192-202. doi: 10.1016/j.riam.2017.02.006. Epub 2017 Jul 18. Rev Iberoam Micol. 2017. PMID: 28732778 Review.
-
Coordinated regulation of arbuscular mycorrhizal fungi and soybean MAPK pathway genes improved mycorrhizal soybean drought tolerance.Mol Plant Microbe Interact. 2015 Apr;28(4):408-19. doi: 10.1094/MPMI-09-14-0251-R. Mol Plant Microbe Interact. 2015. PMID: 25390189
-
Protein-protein interactions in plant mitogen-activated protein kinase cascades.J Exp Bot. 2016 Feb;67(3):607-18. doi: 10.1093/jxb/erv508. Epub 2015 Dec 7. J Exp Bot. 2016. PMID: 26646897 Review.
-
MAP kinases in plant signal transduction: how many, and what for?Results Probl Cell Differ. 2000;27:11-27. doi: 10.1007/978-3-540-49166-8_2. Results Probl Cell Differ. 2000. PMID: 10533195 Review.
Cited by
-
The Mitogen-Activated Protein Kinase PlMAPK2 Is Involved in Zoosporogenesis and Pathogenicity of Peronophythoralitchii.Int J Mol Sci. 2021 Mar 29;22(7):3524. doi: 10.3390/ijms22073524. Int J Mol Sci. 2021. PMID: 33805371 Free PMC article.
-
The mitogen-activated protein kinase module CcSte11-CcSte7-CcPmk1 regulates pathogenicity via the transcription factor CcSte12 in Cytospora chrysosperma.Stress Biol. 2024 Jan 16;4(1):4. doi: 10.1007/s44154-023-00142-w. Stress Biol. 2024. PMID: 38225467 Free PMC article.
-
A complete MAP kinase cascade controls hyphopodium formation and virulence of Verticillium dahliae.aBIOTECH. 2023 May 2;4(2):97-107. doi: 10.1007/s42994-023-00102-y. eCollection 2023 Jun. aBIOTECH. 2023. PMID: 37581020 Free PMC article.
-
Ancient signals: comparative genomics of green plant CDPKs.Trends Plant Sci. 2014 Feb;19(2):79-89. doi: 10.1016/j.tplants.2013.10.009. Epub 2013 Dec 14. Trends Plant Sci. 2014. PMID: 24342084 Free PMC article. Review.
-
Characterisation of HvVIP1 and expression profile analysis of stress response regulators in barley under Agrobacterium and Fusarium infections.PLoS One. 2019 Jun 14;14(6):e0218120. doi: 10.1371/journal.pone.0218120. eCollection 2019. PLoS One. 2019. PMID: 31199821 Free PMC article.
References
-
- Ahn I.P., Suh S.C. (2007). Calcium/calmodulin-dependent signaling for pre-penetration development in Cochliobolus miyabeanus infecting rice. J. Gen. Plant Pathol. 73: 113–120
-
- Andreasson E., Ellis B. (2010). Convergence and specificity in the Arabidopsis MAPK nexus. Trends Plant Sci. 15: 106–113 - PubMed
-
- Andrews D.L., Egan J.D., Mayorga M.E., Gold S.E. (2000). The Ustilago maydis ubc4 and ubc5 genes encode members of a MAP kinase cascade required for filamentous growth. Mol. Plant Microbe Interact. 13: 781–786 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

