Epigenetic regulation of cardiac development and function by polycomb group and trithorax group proteins
- PMID: 22514007
- PMCID: PMC3426356
- DOI: 10.1002/dvdy.23796
Epigenetic regulation of cardiac development and function by polycomb group and trithorax group proteins
Abstract
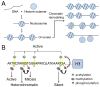
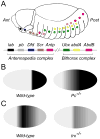
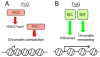
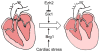
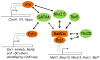
Heart disease is a leading cause of death and disability in developed countries. Heart disease includes a broad range of diseases that affect the development and/or function of the cardiovascular system. Some of these diseases, such as congenital heart defects, are present at birth. Others develop over time and may be influenced by both genetic and environmental factors. Many of the known heart diseases are associated with abnormal expression of genes. Understanding the factors and mechanisms that regulate gene expression in the heart is essential for the detection, treatment, and prevention of heart diseases. Polycomb Group (PcG) and Trithorax Group (TrxG) proteins are special families of chromatin factors that regulate developmental gene expression in many tissues and organs. Accumulating evidence suggests that these proteins are important regulators of development and function of the heart as well. A better understanding of their roles and functional mechanisms will translate into new opportunities for combating heart disease.
Copyright © 2012 Wiley Periodicals, Inc.
Figures


Similar articles
-
Polycomb group protein-associated chromatin is reproduced in post-mitotic G1 phase and is required for S phase progression.J Biol Chem. 2008 Jul 4;283(27):18905-15. doi: 10.1074/jbc.M709322200. Epub 2008 May 2. J Biol Chem. 2008. PMID: 18453536
-
Functional conservation of Asxl2, a murine homolog for the Drosophila enhancer of trithorax and polycomb group gene Asx.PLoS One. 2009;4(3):e4750. doi: 10.1371/journal.pone.0004750. Epub 2009 Mar 9. PLoS One. 2009. PMID: 19270745 Free PMC article.
-
Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins.Nat Rev Cancer. 2010 Oct;10(10):669-82. doi: 10.1038/nrc2931. Nat Rev Cancer. 2010. PMID: 20865010 Free PMC article. Review.
-
Antagonizing Polycomb group-mediated gene repression by chromatin remodelers.Epigenetics. 2010 Jan 1;5(1):20-3. doi: 10.4161/epi.5.1.10559. Epub 2010 Jan 6. Epigenetics. 2010. PMID: 20083900 Review.
-
Critical role of the Polycomb and Trithorax complexes in the maintenance of CD4 T cell memory.Semin Immunol. 2009 Apr;21(2):78-83. doi: 10.1016/j.smim.2009.02.001. Epub 2009 Mar 9. Semin Immunol. 2009. PMID: 19269851 Review.
Cited by
-
Polycomb Group Protein CBX7 Represses Cardiomyocyte Proliferation Through Modulation of the TARDBP/RBM38 Axis.Circulation. 2023 Jun 13;147(24):1823-1842. doi: 10.1161/CIRCULATIONAHA.122.061131. Epub 2023 May 9. Circulation. 2023. PMID: 37158107 Free PMC article.
-
A role for BRG1 in the regulation of genes required for development of the lymphatic system.Oncotarget. 2017 Jul 4;8(33):54925-54938. doi: 10.18632/oncotarget.18976. eCollection 2017 Aug 15. Oncotarget. 2017. PMID: 28903392 Free PMC article.
-
Epigenetic regulation in heart failure.Curr Opin Cardiol. 2016 May;31(3):255-65. doi: 10.1097/HCO.0000000000000276. Curr Opin Cardiol. 2016. PMID: 27022893 Free PMC article. Review.
-
Epigenetic modifications: basic mechanisms and role in cardiovascular disease (2013 Grover Conference series).Pulm Circ. 2014 Jun;4(2):169-74. doi: 10.1086/675979. Pulm Circ. 2014. PMID: 25006435 Free PMC article. Review.
-
Asxl2-/- Mice Exhibit De Novo Cardiomyocyte Production during Adulthood.J Dev Biol. 2016 Nov 3;4(4):32. doi: 10.3390/jdb4040032. J Dev Biol. 2016. PMID: 29615595 Free PMC article.
References
-
- Aalfs JD, Kingston RE. What does “chromatin remodeling” mean? Trends in biochemical sciences. 2000;25:548–555. - PubMed
-
- Balza RO, Jr, Misra RP. Role of the serum response factor in regulating contractile apparatus gene expression and sarcomeric integrity in cardiomyocytes. J Biol Chem. 2006;281:6498–6510. - PubMed
-
- Berger SL. Histone modifications in transcriptional regulation. Current opinion in genetics & development. 2002;12:142–148. - PubMed
-
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, Schreiber SL, Lander ES. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

