Practical structure solution with ARCIMBOLDO
- PMID: 22505254
- PMCID: PMC3322593
- DOI: 10.1107/S0907444911056071
Practical structure solution with ARCIMBOLDO
Abstract
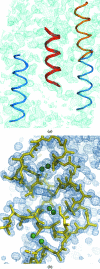
Since its release in September 2009, the structure-solution program ARCIMBOLDO, based on the combination of locating small model fragments such as polyalanine α-helices with density modification with the program SHELXE in a multisolution frame, has evolved to incorporate other sources of stereochemical or experimental information. Fragments that are more sophisticated than the ubiquitous main-chain α-helix can be proposed by modelling side chains onto the main chain or extracted from low-homology models, as locally their structure may be similar enough to the unknown one even if the conventional molecular-replacement approach has been unsuccessful. In such cases, the program may test a set of alternative models in parallel against a specified figure of merit and proceed with the selected one(s). Experimental information can be incorporated in three ways: searching within ARCIMBOLDO for an anomalous fragment against anomalous differences or MAD data or finding model fragments when an anomalous substructure has been determined with another program such as SHELXD or is subsequently located in the anomalous Fourier map calculated from the partial fragment phases. Both sources of information may be combined in the expansion process. In all these cases the key is to control the workflow to maximize the chances of success whilst avoiding the creation of an intractable number of parallel processes. A GUI has been implemented to aid the setup of suitable strategies within the various typical scenarios. In the present work, the practical application of ARCIMBOLDO within each of these scenarios is described through the distributed test cases.
Figures

Similar articles
-
Macromolecular ab initio phasing enforcing secondary and tertiary structure.IUCrJ. 2015 Jan 1;2(Pt 1):95-105. doi: 10.1107/S2052252514024117. eCollection 2015 Jan 1. IUCrJ. 2015. PMID: 25610631 Free PMC article. Review.
-
Structure solution with ARCIMBOLDO using fragments derived from distant homology models.FEBS J. 2014 Sep;281(18):4029-45. doi: 10.1111/febs.12897. Epub 2014 Sep 6. FEBS J. 2014. PMID: 24976038
-
SEQUENCE SLIDER: expanding polyalanine fragments for phasing with multiple side-chain hypotheses.Acta Crystallogr D Struct Biol. 2020 Mar 1;76(Pt 3):221-237. doi: 10.1107/S2059798320000339. Epub 2020 Feb 25. Acta Crystallogr D Struct Biol. 2020. PMID: 32133987 Free PMC article.
-
ALIXE: a phase-combination tool for fragment-based molecular replacement.Acta Crystallogr D Struct Biol. 2020 Mar 1;76(Pt 3):209-220. doi: 10.1107/S205979832000056X. Epub 2020 Feb 25. Acta Crystallogr D Struct Biol. 2020. PMID: 32133986 Free PMC article.
-
Experimental Phasing: Substructure Solution and Density Modification as Implemented in SHELX.Methods Mol Biol. 2017;1607:357-376. doi: 10.1007/978-1-4939-7000-1_15. Methods Mol Biol. 2017. PMID: 28573581 Review.
Cited by
-
Macromolecular ab initio phasing enforcing secondary and tertiary structure.IUCrJ. 2015 Jan 1;2(Pt 1):95-105. doi: 10.1107/S2052252514024117. eCollection 2015 Jan 1. IUCrJ. 2015. PMID: 25610631 Free PMC article. Review.
-
Structure and cell wall cleavage by modular lytic transglycosylase MltC of Escherichia coli.ACS Chem Biol. 2014 Sep 19;9(9):2058-66. doi: 10.1021/cb500439c. Epub 2014 Jul 10. ACS Chem Biol. 2014. PMID: 24988330 Free PMC article.
-
Fragment-based determination of a proteinase K structure from MicroED data using ARCIMBOLDO_SHREDDER.Acta Crystallogr D Struct Biol. 2020 Aug 1;76(Pt 8):703-712. doi: 10.1107/S2059798320008049. Epub 2020 Jul 27. Acta Crystallogr D Struct Biol. 2020. PMID: 32744252 Free PMC article.
-
Extending molecular-replacement solutions with SHELXE.Acta Crystallogr D Biol Crystallogr. 2013 Nov;69(Pt 11):2251-6. doi: 10.1107/S0907444913027534. Epub 2013 Oct 18. Acta Crystallogr D Biol Crystallogr. 2013. PMID: 24189237 Free PMC article.
-
Structural characterization of KKT4, an unconventional microtubule-binding kinetochore protein.Structure. 2021 Sep 2;29(9):1014-1028.e8. doi: 10.1016/j.str.2021.04.004. Epub 2021 Apr 28. Structure. 2021. PMID: 33915106 Free PMC article.
References
-
- Bieniossek, C., Schütz, P., Bumann, M., Limacher, A., Usón, I. & Baumann, U. (2006). J. Mol. Biol. 360, 457–465. - PubMed
-
- Caliandro, R., Carrozzini, B., Cascarano, G. L., De Caro, L., Giacovazzo, C., Mazzone, A. & Siliqi, D. (2008). J. Appl. Cryst. 41, 548–553.
-
- Caliandro, R., Carrozzini, B., Cascarano, G. L., De Caro, L., Giacovazzo, C. & Siliqi, D. (2005a). Acta Cryst. D61, 556–565. - PubMed
-
- Caliandro, R., Carrozzini, B., Cascarano, G. L., De Caro, L., Giacovazzo, C. & Siliqi, D. (2005b). Acta Cryst. D61, 1080–1087. - PubMed
-
- DeLano, W. L. (2002). PyMOL http://www.pymol.org.

